Authors:
Li Pan、Yupei Zhang、Qiushi Yang、Tan Li、Xiaohan Xing、Maximus C. F. Yeung、Zhen Chen
Paper:
https://arxiv.org/abs/2408.08527
Introduction
Gliomas, the most common type of brain tumors, are classified into Grades II to IV by the World Health Organization (WHO), with each grade correlating with different prognoses and intervention approaches. The gold standard for grading gliomas involves the observation of representative histopathology features in biopsies. However, histopathology slides present a complex milieu of cells, necrosis, and microenvironments, complicating the localization of tumor foci and necessitating the expertise of senior pathologists.
Recent advances in computer-assisted cancer grading have shown promising performance in identifying glioma grades from histopathology slides. Multimodal approaches that combine histopathology features with molecular biomarkers from tissue biopsies offer a comprehensive and accurate tumor analysis. However, these methods face challenges due to the intra-modality complexity and inter-modality heterogeneity, leading to inadequate histopathology representation learning and inefficient molecular-pathology knowledge alignment.
To address these challenges, the authors propose a novel Focus on Focus (FoF) framework with paired pathology-genomic training and applicable pathology-only inference, enhancing molecular-pathology representation effectively.
Related Work
Glioma Grading
Histopathology grading of gliomas involves the classification of malignant tumors occurring in the glial cells of the brain or spinal cord into Grades II to IV. Traditional machine learning and deep learning models have been employed to classify morphological features observed under a microscope from biopsy samples. However, these approaches often lack generalization ability and interpretability due to the high complexity of histopathology slides.
Multimodal Glioma Grading with Missing Modality
Molecular biomarkers are essential for the clinical assessment of cancers, but their acquisition requires immunohistochemistry (IHC) staining and DNA/RNA sequencing, which are time-consuming and costly. Fusion-based multimodal methods necessitate the presence of paired histopathology slides and molecular biomarkers, further restricting their practical application in real-world clinical settings. Recent studies have implemented distillation-based methods to enhance image-based models by distilling knowledge from a pathology-genomic teacher, achieving performance comparable to multimodal grading using only histopathology slides.
Methodology
Overview
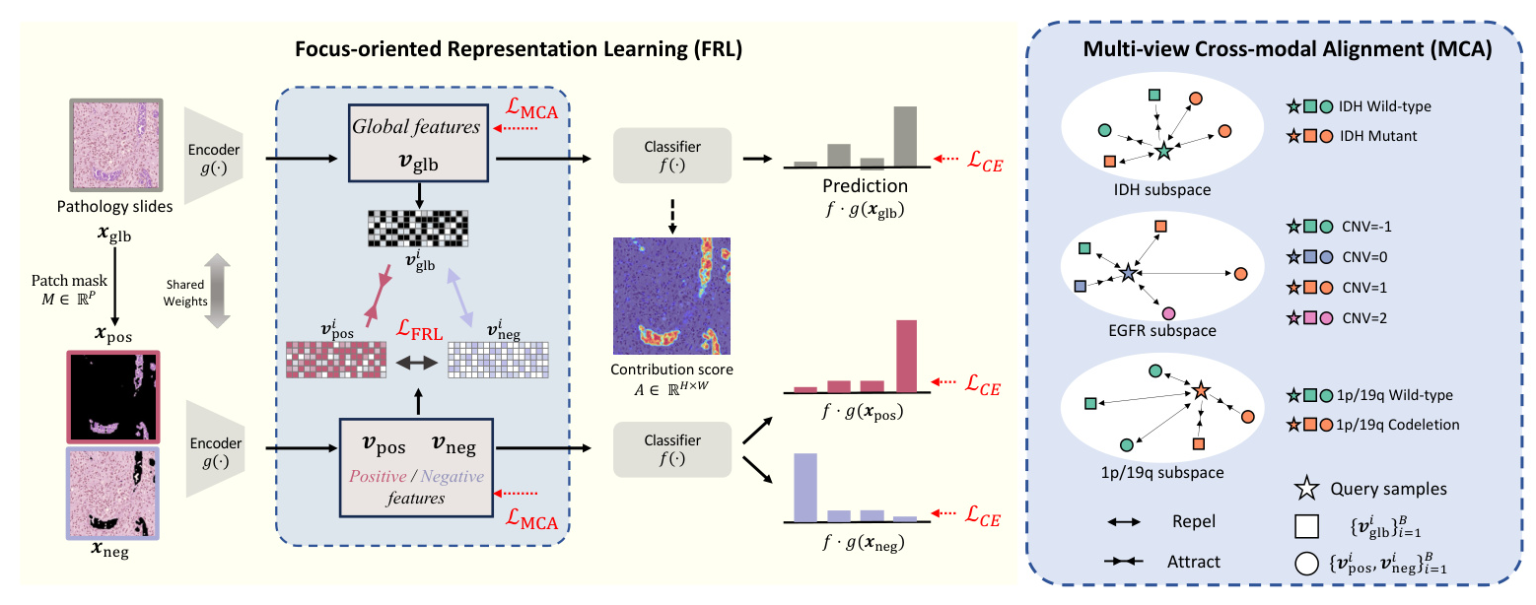
The FoF framework improves glioma grading by highlighting diagnostic molecular-pathology features. It introduces Focus-oriented Representation Learning (FRL) to identify regions positively and negatively correlated to cancer grading and Multi-view Cross-modal Alignment (MCA) to project histopathology representations into molecular subspaces, aligning morphological features with corresponding molecular biomarker status through supervised contrastive learning.
Focus-oriented Representation Learning
FRL quantifies the contribution score of each pixel towards accurate classification and identifies areas positively and negatively related to grading using a threshold. This module enhances visual representations with a consistency constraint on the positive and global features.
Technically, FRL processes the whole image through the model to obtain predictions and aggregates the gradient of the prediction for the ground-truth class across each feature map layer, estimating the contribution score of each pixel. The pixel-wise contribution score is divided into patches, and a mask is generated to select positive and negative patches. The Cross-Entropy loss is applied to all regions, and a consistency constraint promotes a consistent mapping of positive and overall regions while distinguishing negative regions.
Multi-view Cross-modal Alignment
MCA employs genomic biomarkers as unique labels and aligns histological feature representations within each molecular subspace by supervised contrastive learning. It leverages biomarkers with discrete values, such as IDH mutation status and 1p/19q codeletion presence, as labels. This module projects multi-view histopathology representations into individual molecular subspaces, bringing features that share the same molecular biomarker values closer together while distancing those that differ.
Training and Inference
The overall optimization objective of the FoF framework combines the losses from FRL and MCA modules. During training, the framework estimates the contribution score in the first back-propagation and calculates the losses in the second back-propagation. During inference, the model directly outputs predictions from input images with no additional computational cost.
Experiment
Dataset and Implementation Details
The FoF framework is evaluated on the TCGA-GBM and TCGA-LGG datasets, comprising paired histopathology slides and genomic profiles. The dataset includes 736 patients with standard grading labels. The ROI images are augmented through random cropping, color jittering, flipping, and distortion. The framework is implemented with the PyTorch library and employs ViT-Tiny as the encoder for images. The experiments are conducted on four NVIDIA GeForce GTX 1080 Ti GPUs with a batch size of 4.
Comparison with Histopathology Grading Methods
The FoF framework is compared with state-of-the-art methods that require only histopathology slides as input during inference. The results show that FoF outperforms other histopathology grading methods in all metrics on the TCGA GBM-LGG dataset, achieving higher accuracy, AUC, average precision, and Kappa scores.
Comparison with Multimodal Grading Methods
The FoF framework is also compared with multimodal grading methods that require paired histopathology slides and genomic data. The results demonstrate that FoF with sole histopathology slides outperforms all multimodal grading methods on all metrics, illustrating the effectiveness of the proposed framework.
Qualitative Analysis
The distributions of morphological features regarding different values of molecular biomarkers are illustrated, showing a clearer clustering of histopathology features with respect to molecular biomarkers in the FoF framework compared to other methods. The model focus on input histopathology slides is also compared, with FoF concentrating on critical indicators for diagnosing high-grade gliomas.
Conclusion
The FoF framework utilizes pathology-genomic knowledge for accurate glioma grading with histopathology slides. The FRL module encourages the model to focus on diagnostic regions, and the MCA scheme efficiently aligns molecular biomarkers with visual representations. Experimental results indicate that FoF significantly improves glioma grading performance, achieving superior results with sole histopathology slides compared to existing multimodal approaches, demonstrating great clinical significance.