Authors:
Junlin Guo、Siqi Lu、Can Cui、Ruining Deng、Tianyuan Yao、Zhewen Tao、Yizhe Lin、Marilyn Lionts、Quan Liu、Juming Xiong、Catie Chang、Mitchell Wilkes、Mengmeng Yin、Haichun Yang、Yuankai Huo
Paper:
https://arxiv.org/abs/2408.06381
Introduction
Cell nuclei instance segmentation is a fundamental task in digital pathology, particularly for accurate disease diagnosis and treatment planning. However, the generalizability of current methodologies to handle diverse and large-scale datasets remains a significant challenge. This study evaluates the performance of state-of-the-art (SOTA) cell nuclei foundation models in kidney pathology, focusing on three widely used models: Cellpose, StarDist, and CellViT.
Methods
Diverse Large-scale Dataset
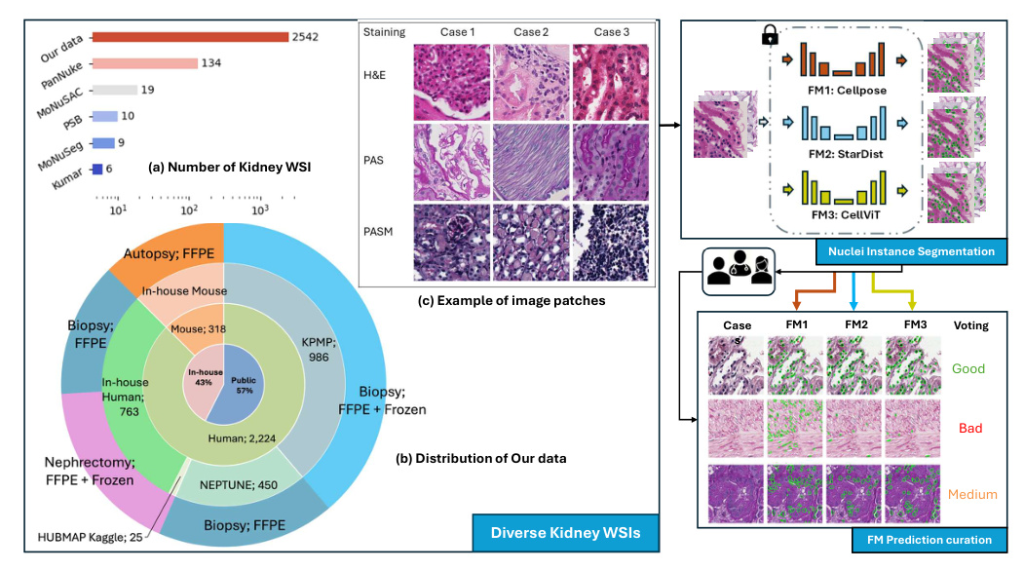
A diverse evaluation dataset was created, consisting of 2,542 kidney whole slide images (WSIs) from both human and rodent sources. These WSIs were collected from public datasets such as Kidney Tissue Atlas (KPMP), NEPTUNE, and HUBMAP, as well as an in-house collection at Vanderbilt University Medical Center. The dataset includes various tissue types and staining methods, such as Hematoxylin and Eosin (H&E), Periodic acid-Schiff methenamine (PASM), and Periodic acid-Schiff (PAS).
Nuclei Instance Segmentation
Nuclei instance segmentation was performed using three foundation models: Cellpose, StarDist, and CellViT. Each model has unique features and methodologies:
- Cellpose: Utilizes a U-Net backbone to predict topological maps and a binary map, retrieving individual objects through a gradient tracking algorithm.
- StarDist: Employs a star-convex polygon representation for nuclei segmentation, using a U-Net architecture for dense prediction of radial distances and object probabilities.
- CellViT: Uses a hierarchical encoder-decoder Vision Transformer (ViT) backbone, leveraging pre-trained weights from a ViT trained on 104 million histological images.
Rating on Segmentation Performance
Two pathologist-trained students scored the model predictions as “good,” “medium,” or “bad” based on a standard provided by a renal pathologist. This scoring system quantitatively assessed each model’s predictions, and the ratings were used to curate strongly agreed-upon failure samples for future model training or fine-tuning.
Experiments
Distribution of Foundation Model Predictions
The distribution of predictions for each foundation model was analyzed across the evaluation dataset. This analysis provided insights into the performance and behavior of each model by examining the frequency of different prediction classes (“good,” “medium,” “bad”).
Agreement Analysis Between Foundation Models
An agreement analysis was conducted to assess cross-model performance on the kidney nuclei dataset. This involved calculating agreement percentages between each pair of models and categorizing image patches based on the level of agreement among the models.
Consensus “Bad” Image Patches Mining
By curating labels from the three foundation models, strong-agreed-upon failure samples were identified. These “global failure” image patches highlight areas where current cell nuclei foundation models need improvement in kidney pathology.
Results
Distribution of Predictions
The distribution of predictions for each model revealed that at least 40% of predictions were categorized as “bad” or “medium,” while at least 40% were “good.” This indicates that the models retain some transferable knowledge applicable to kidney pathology, but a performance gap remains.
Agreement Analysis
The agreement matrix showed that no pair of models exhibited over 90% agreement or complete disagreement. The highest agreement was observed between Cellpose and StarDist (0.76), while the lowest was between Cellpose and CellViT (0.67). This inconsistency suggests that combining multiple SOTA foundation models could enhance performance in downstream kidney nuclei tasks.
Consensus “Good” and “Failure” Examples
Consensus “good” examples showed that current nuclei foundation models perform well on image patches with darker nuclei and higher contrast. Conversely, consensus “failure” examples highlighted challenges with lower staining intensity, lighter nuclei, and reduced imaging contrast.
Conclusion
This study provides a comprehensive assessment of cell nuclei foundation models (Cellpose, StarDist, CellViT) in kidney pathology, focusing on nuclei instance segmentation. CellViT demonstrated superior performance, but a performance gap remains between general and kidney-specific nuclei segmentation. The curated consensus “good” and “failure” image patches offer valuable insights for improving cell nuclei foundation models in kidney pathology.
Acknowledgments
This work was supported by the National Institutes of Health, the National Science Foundation, and the Vanderbilt Institute for Clinical and Translational Research. Additional support was provided by NVIDIA through a hardware grant.