Authors:
Xiaohan Cheng、Taiyuan Mei、Yun Zi、Qi Wang、Zijun Gao、Haowei Yang
Paper:
https://arxiv.org/abs/2408.06357
Introduction
Medical image recognition technology, leveraging deep learning, has made significant strides. Traditional algorithms, however, require extensive labeled samples and struggle with novel categories. Zero-shot learning (ZSL) addresses this by enabling models to recognize unseen categories. This research explores embedding-based ZSL methods, focusing on aligning features and quasi-semantic information of medical images within a vector space. The challenge lies in avoiding overfitting known categories and ensuring accurate predictions for unknown categories. The study shifts from convolutional neural networks to Transformer-based frameworks, aiming to improve model performance through semantic similarity-based multi-label category loss and ELMo-MCT.
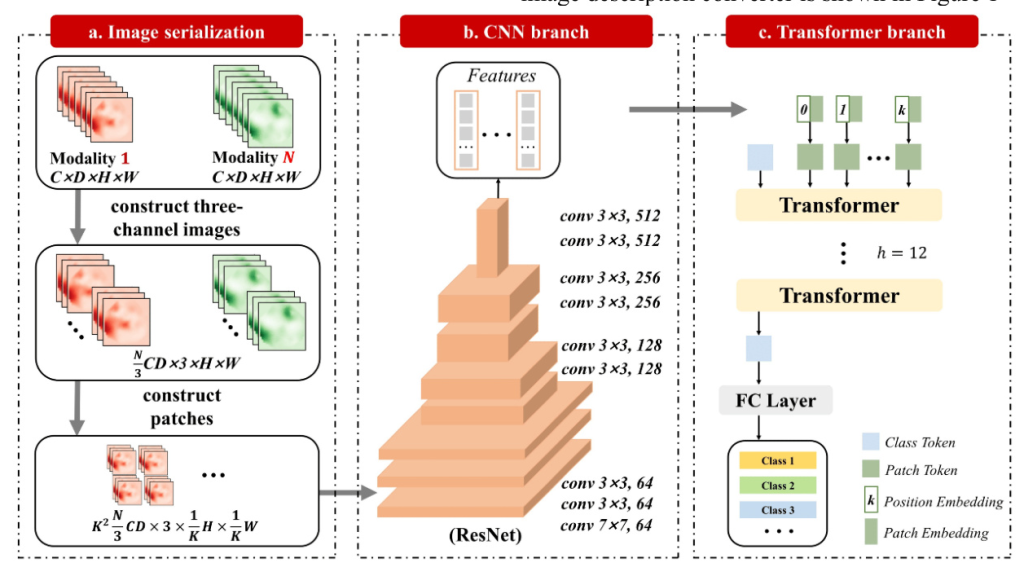
Multi-Mode Image Description Converter
Image Feature Encoder
The core task of the image feature encoder is to extract image characteristics and establish an attention matrix between image modes using an attention mechanism. The process involves two main components:
- Image Feature Extractor: Utilizing a bottom-up attention mechanism, the Faster-RCNN identifies object association regions. Mean pooling is applied to obtain features for each object, forming an image feature matrix consistent with the encoder’s spatial scale.
- Multi-Channel Converter: The converted image feature is input to a multimode converter with multiple attention modules. Each module computes attention characteristics in the image mode, with the multi-head attention mechanism playing a crucial role.
Text Decoder
The text decoder translates the image characteristic matrix into text. It involves:
- Multi-Mode Lexical Information Hidden Encoder: This encoder constructs character representations by encoding input words. It uses both standard word embedding and ELMo word embedding encoders to generate feature matrices.
- Multimodal Transformer Decoder: This module models the input word embedding matrix using masked multi-head attention, generating attention characteristic matrices for both word and image patterns.
Experimental Evaluation
Data Set
The study utilizes the Microsoft COCO2014 open-source database, dividing samples into validation, training, and testing sets. Evaluation metrics include BLEU, ROUGE-L, and CIDErD, with the dataset managed using Linked Data methodology to enhance interoperability.
Experimental Details
Each image is paired with five captions in English, converted to lower case and tokenized. The model parameters include feature dimensions, head quantities, and term vectors. Training involves 30 cycles with cross-loss method and Adam optimizer.
Experimental Results and Analysis
The study compares image characteristics and LSTM-based models with MCT and ELMo-MCT. Results show significant improvements in CIDEr scores for ELMo-MCT, indicating richer semantic meaning in word vector coding. The effectiveness of the method decreases with increased depth, suggesting potential information loss over time.
Conclusion
The research highlights the limitations of traditional word embedding methods like Word2Vec and Glove, which result in data loss. By establishing a multi-mode converter and integrating ELMo with vector methods, the model’s semantic expression level is enhanced. The findings demonstrate the potential of combining semantic similarity measures with advanced feature extraction technologies to improve zero-shot learning in medical image recognition. This approach not only boosts model performance but also reduces dependency on extensive labeled datasets, paving the way for more adaptable and efficient medical imaging technologies.