Authors:
Gregory Kell、Angus Roberts、Serge Umansky、Yuti Khare、Najma Ahmed、Nikhil Patel、Chloe Simela、Jack Coumbe、Julian Rozario、Ryan-Rhys Griffiths、Iain J. Marshall
Paper:
https://arxiv.org/abs/2408.08624
RealMedQA: A Pilot Biomedical Question Answering Dataset Containing Realistic Clinical Questions
Introduction
Clinical question answering (QA) systems have the potential to revolutionize the way clinicians access information during consultations. Despite significant advancements, the adoption of these systems in clinical settings remains limited. One major barrier is the lack of QA datasets that reflect the real-world needs of healthcare professionals. Existing datasets often derive questions from research article titles or are designed to test comprehension of biomedical corpora, which may not align with the practical needs of clinicians.
To address this gap, the RealMedQA dataset was developed. This dataset consists of realistic clinical questions generated by both medical students and a large language model (LLM), with answers verified by medical professionals. The dataset aims to provide reliable, guidance-oriented, and rationale-backed answers to clinical questions, thereby enhancing the utility of QA systems in clinical practice.
Methods
The methodology for creating the RealMedQA dataset involved three main stages: data collection, question generation, and question-answer pair verification.
Data Collection
Guidelines from the UK National Institute for Health and Care Excellence (NICE) were downloaded via the NICE syndication API. The focus was on guidelines related to care provision, resulting in a total of 7,385 guidelines.
Question Generation
Six UK-based medical students were hired to generate questions based on the NICE guidelines. Each student was asked to create up to five questions per recommendation, focusing on questions that clinicians would likely ask during practice. Additionally, the LLM (gpt-3.5-turbo) was used to generate up to 20 questions per recommendation. In total, 1150 QA pairs were generated by humans and 100,325 by the LLM.
Question-Answer Pair Verification
The QA pairs underwent a verification process by four of the medical students. Each QA pair was evaluated for plausibility and whether the question was answered by the guideline using a Likert scale. The verification process aimed to identify “ideal” QA pairs, where both the question and answer were deemed completely plausible and completely answered.
Experiments
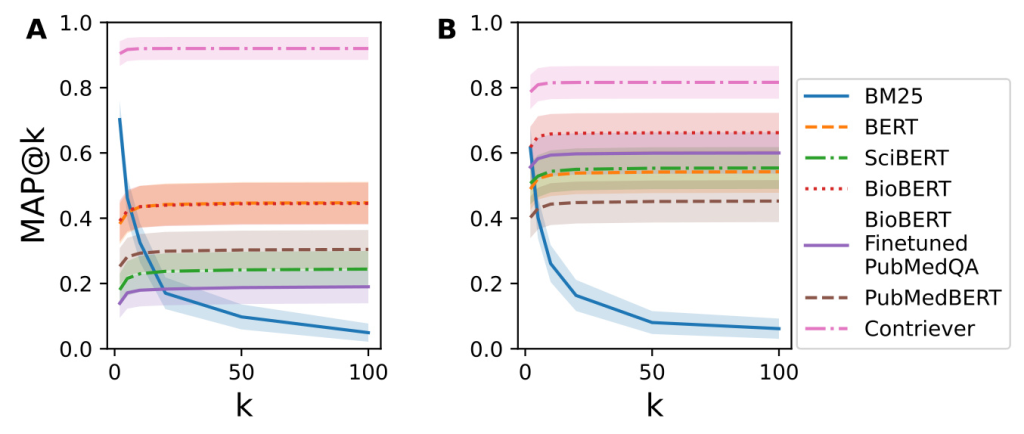
To assess the difficulty of matching RealMedQA’s questions and answers, several QA models were evaluated using metrics such as recall@k, nDCG@k, and MAP@k. The models included BM25, BERT, SciBERT, BioBERT, PubMedBERT, and Contriever. The performance of these models on RealMedQA was compared with their performance on the BioASQ dataset.
Results
Plausibility and Answerability
Out of 1200 QA pairs, only 230 (19%) were deemed “ideal” (completely plausible and completely answered)
Quantitative Comparison of BioASQ and RealMedQA
The recall@k and MAP@k metrics showed that BM25 performed better on BioASQ for lower values of k, while Contriever outperformed BM25 on RealMedQA
Qualitative Comparison of RealMedQA and BioASQ
A qualitative comparison revealed that BioASQ questions had higher lexical similarity with their answers compared to RealMedQA. This difference highlights the more practical and clinically relevant nature of RealMedQA questions.
Discussion
LLM vs Human QA Pair Quality
The yield of “ideal” QA pairs was 7.4% higher for human-generated questions compared to those generated by the LLM. However, the cost-efficiency of LLMs makes them a competitive option, especially when considering the cost per QA pair.
LLM vs Human QA Pair Costs
The cost of generating “ideal” QA pairs using LLMs was significantly lower than using human generators, even after accounting for verification costs. This cost-efficiency could be further improved with automatic verification methods.
Comparison with Other Biomedical QA Datasets
RealMedQA addresses a unique gap by focusing on realistic clinical questions and verified guideline-based answers. This sets it apart from other biomedical QA datasets that often emphasize comprehension of biomedical literature.
Limitations
The study acknowledges the rapid evolution of generative AI and the potential impact of newer models on cost and performance. Additionally, the dataset is currently too small for training purposes, but plans are in place to expand it using LLMs and medical professional verification.
Conclusion
RealMedQA presents a promising methodology for creating biomedical QA datasets with realistic clinical questions and verified answers. The dataset demonstrates the potential of LLMs in generating cost-efficient QA pairs, although human verification remains crucial for ensuring quality. The findings suggest that RealMedQA could drive methodological advancements in biomedical QA systems, ultimately improving their utility in clinical practice.
References
- https://github.com/gck25/RealMedQA
- https://huggingface.co/datasets/k2141255/RealMedQA