Authors:
Jiaojiao Guan、Yongxin Ji、Cheng Peng、Wei Zou、Xubo Tang、Jiayu Shang、Yanni Sun
Paper:
https://arxiv.org/abs/2408.06402
PhaGO: Protein Function Annotation for Bacteriophages by Integrating Genomic Context
Introduction
Bacteriophages, or phages, are viruses that infect bacterial cells and are abundant in various environments such as animal gastrointestinal tracts, water bodies, and soil. They play a crucial role in microbial ecology by influencing bacterial adaptation, evolution, and population dynamics. Due to the increasing threat of antibiotic resistance, phages are gaining attention as potential alternatives to traditional antibiotics.
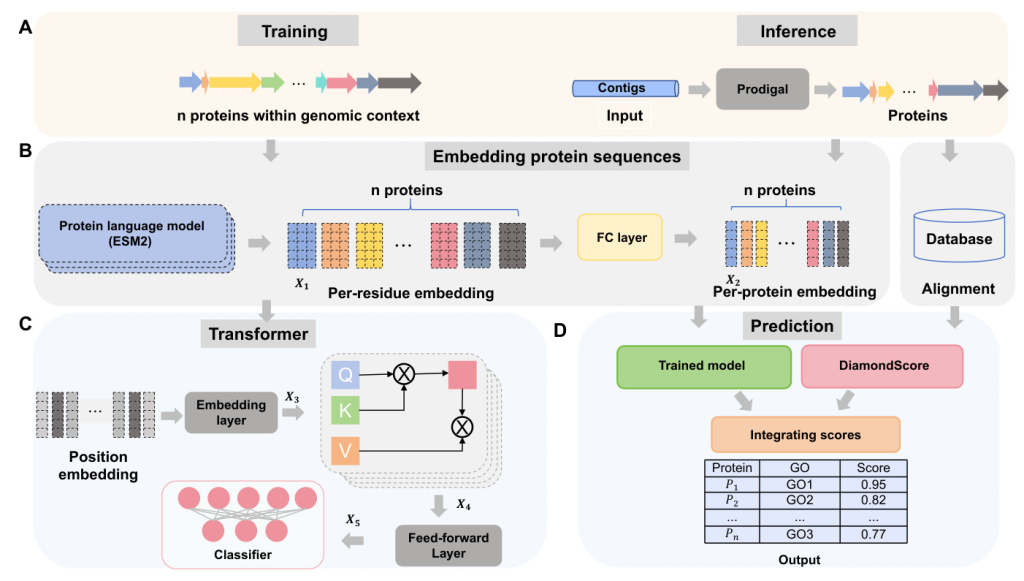
Understanding phage biology, including virus infection, replication, and evolution, requires accurate annotation of phage proteins. However, the diversity and scarcity of annotated phage proteins present significant challenges. Existing tools have not fully leveraged the unique properties of phages for protein function annotation. This paper introduces PhaGO, a new tool that integrates the modular genomic structure of phage genomes with embeddings from the latest protein foundation models and Transformer to improve protein function annotation.
Methods and Materials
Embedding Protein Sequences
The PhaGO model uses the ESM2 protein language model to encode phage proteins. ESM2 is pre-trained on protein sequences and can effectively generate meaningful representations for phage proteins. The model processes protein sequences to generate an embedding matrix, which is then passed through a fully connected layer to produce a one-dimensional feature set.
Learning the Relationship of Context Proteins Using Transformer
Proteins within the same genus of phages tend to maintain a consistent arrangement, similar to words in a sentence. PhaGO leverages this contextual information by treating each protein as a token and contigs as sentences. The model uses position embedding to encode the relative position of each protein within the sequence. A self-attention layer learns the intricate connections between proteins, and a multi-head mechanism focuses on diverse representation subspaces.
Predicting the GO Terms
Protein function annotation is formulated as a multi-label binary classification task. The model assigns a probability to each Gene Ontology (GO) term, indicating the likelihood of the protein being associated with that specific function. The model is optimized by minimizing the binary cross-entropy loss.
Integrating PhaGO with Alignment-Based Method
PhaGO+ combines PhaGO with DiamondScore, an alignment-based method, to enhance predictive capabilities. The confidence scores from both methods are integrated to improve the accuracy of protein function predictions.
Results
Metrics
The performance of PhaGO is evaluated using protein-centric and GO term-centric metrics. Protein-centric evaluation focuses on function prediction accuracy, while term-centric evaluation examines the model’s ability to identify proteins associated with specific functional terms.
Dataset
The dataset includes reference genomes and proteins from the Caudoviricetes class, annotated using the UniProt database and the Prokaryotic Virus Remote Homologous Groups (PHROG) database. Proteins from genera with high annotation rates are selected for training and evaluation.
PhaGO Outperforms State-of-the-Art Predictors
PhaGO+ outperforms existing methods, achieving higher AUPR and Fmax scores across all three GO categories (BP, CC, and MF). The integration of DiamondScore with PhaGO further improves performance, especially for proteins with no alignment to the training dataset.
PhaGO Improves Annotation of Proteins by Utilizing Contextual Information
Experiments show that incorporating contextual proteins significantly enhances function prediction accuracy. The model’s performance improves as the number of context proteins increases, highlighting the importance of considering contextual information.
PhaGO Shows Superior Performance in Annotating Remote Homologous Proteins
PhaGO+ demonstrates robust performance in predicting protein functions for low-similarity proteins, achieving significant improvements in AUPR for proteins with no alignment to the training dataset.
PhaGO Enhances Annotation with Focus on Minority Class GO Terms
PhaGO+ excels in predicting minority GO terms, achieving high AUPR for infrequently occurring terms. The incorporation of context proteins further improves performance for these minority terms.
PhaGO Enables Protein Function Annotation Without Relying on Homology Search
PhaGO successfully annotates holin proteins, a diverse group of small membrane proteins crucial for lysing bacterial hosts. The model identifies 688 potential holin proteins, many of which exhibit structural homology with known holins despite low sequence similarity.
Conclusion and Discussion
PhaGO leverages the unique properties of phages and the power of foundation models to achieve superior protein function annotation. The integration of contextual information and alignment-based methods enhances the model’s performance, particularly for low-similarity and minority GO term labels. PhaGO’s ability to annotate uncharacterized proteins makes it a valuable tool for biological discovery and in-depth investigations.
Future work will focus on incorporating additional features such as structural information and textual descriptions to further improve the annotation process.
Data Availability
PhaGO is implemented in Python and can be downloaded from https://github.com/jiaojiaoguan/PhaGO.
Supplementary Data
Supplementary data are available at NAR Online.
Competing Interests
No competing interests are declared.
Acknowledgments
The authors thank the anonymous reviewers for their valuable suggestions. This work is supported by GRF 11209823, City University of Hong Kong 7005866, and ARG 9667256.