Authors:
Silvia Seidlitz、Katharina Hölzl、Ayca von Garrel、Jan Sellner、Stephan Katzenschlager、Tobias Hölle、Dania Fischer、Maik von der Forst、Felix C.F. Schmitt、Markus A. Weigand、Lena Maier-Hein、Maximilian Dietrich
Paper:
https://arxiv.org/abs/2408.09873
New Spectral Imaging Biomarkers for Sepsis and Mortality in Intensive Care
Introduction
Sepsis is a life-threatening condition characterized by organ dysfunction due to a dysregulated response to infection. It remains a leading cause of mortality worldwide, accounting for nearly 19.7% of global deaths in 2017. Early identification of sepsis and high-risk patients is crucial for improving outcomes, but current diagnostic methods often identify sepsis only at advanced stages, leading to delays in treatment and increased mortality. This study explores the potential of hyperspectral imaging (HSI) as a novel, non-invasive biomarker for early sepsis diagnosis and mortality prediction in intensive care unit (ICU) patients.
Related Work
Existing Biomarkers and Machine Learning Models
Over 250 molecules have been proposed as potential biomarkers for sepsis diagnosis and mortality prediction, but none have demonstrated outstanding sensitivity and specificity. Recent studies have employed machine learning models, particularly random forests, to predict sepsis using high-dimensional clinical data from electronic health records (EHRs). However, these models face challenges such as data standardization, completeness, and inherent biases, limiting their generalizability, especially in low- and middle-income countries (LMICs).
Microcirculatory Monitoring
Monitoring microcirculatory alterations has gained attention due to the decoupling of microcirculation and systemic hemodynamics in sepsis. Imaging methods like sublingual microscopy, laser Doppler flowmetry, and HSI have revealed microcirculatory dysfunction in sepsis, characterized by reduced capillary density and increased heterogeneity in microperfusion. HSI, in particular, offers mobile, non-invasive, rapid, and cost-effective assessments, making it a promising tool for clinical use.
Research Methodology
Hypothesis and Objectives
The study hypothesizes that HSI can serve as an imaging biomarker for automated sepsis diagnosis and mortality prediction by monitoring microcirculatory dysfunction and edema formation. The key research questions are:
1. Can deep learning-based HSI analysis provide automated, non-invasive, and rapid diagnosis of sepsis and prediction of mortality in ICU patients?
2. Can the diagnostic and predictive performance be enhanced by incorporating structured clinical data?
3. Does the proposed method outperform widely used clinical scores and biomarkers?
Data Collection
HSI data and corresponding red-green-blue (RGB) images were collected from the palm and fingers of over 480 ICU patients at the University Hospital Heidelberg. Clinical data, including demographics, vital signs, blood gas analysis, therapy details, and laboratory results, were also recorded.
Experimental Design
Hyperspectral Image Acquisition
The medical device-graded HSI system Tivita® 2.0 Surgery Edition was used to collect HSI data. The system captures tissue reflectance spectra, allowing the approximation of functional parameters like oxygen saturation, perfusion, hemoglobin content, and water content. Measurements were taken from the palm and annular finger due to their easy accessibility and low melanin content.
Data Preprocessing and Annotation
HSI cubes were calibrated using white and dark reference cubes, followed by ℓ1-normalization across spectral channels. Images were cropped and rescaled to standard dimensions, with annotated skin areas used for analysis to avoid confounding elements like dressings or tubes.
Classification Models
Deep learning classifiers, specifically ResNet14d architectures with pre-trained ImageNet weights, were used for HSI and HSI + clinical data models. A random forest classifier was employed for clinical data alone. The models were trained using cross-entropy loss, with data augmentations and regularization techniques applied to improve performance.
Results and Analysis
Diagnostic and Predictive Performance
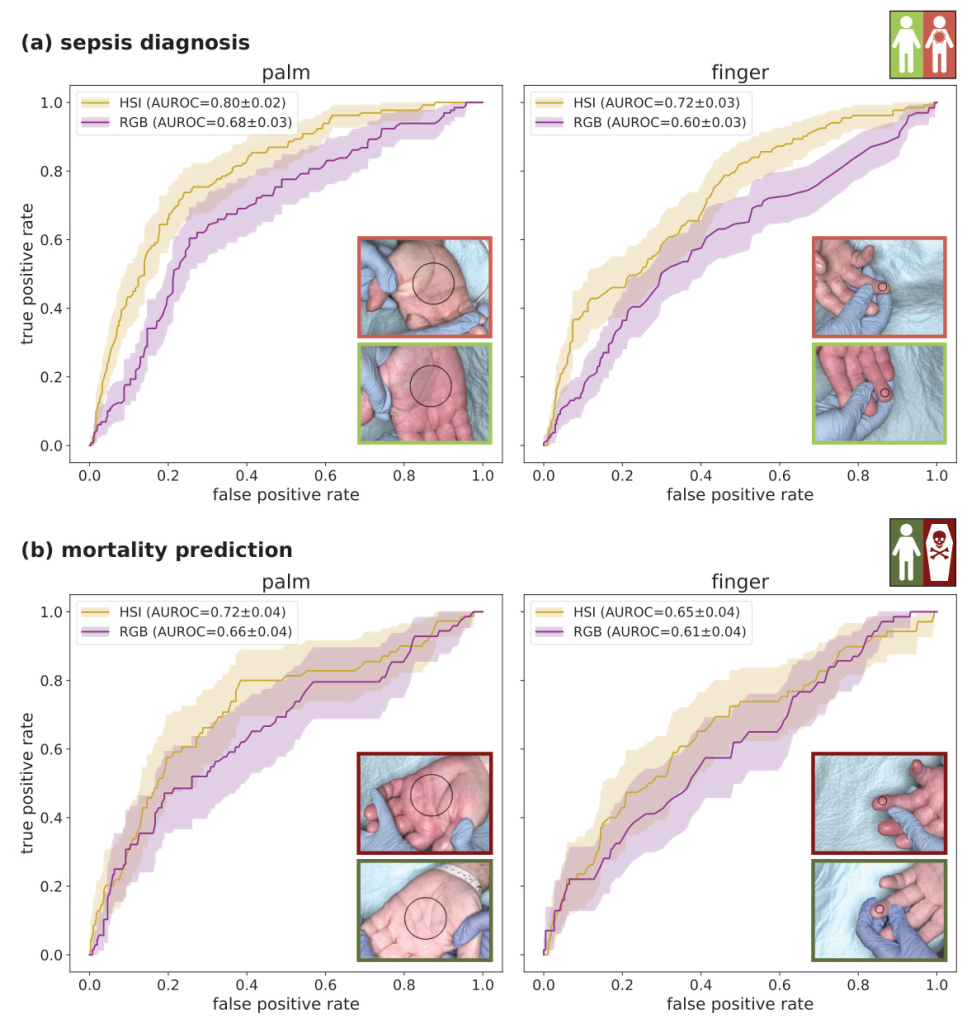
HSI measurements from the palm achieved an AUROC of 0.80 for sepsis diagnosis and 0.72 for mortality prediction, outperforming RGB imaging by up to 20%. Incorporating clinical data improved the AUROC to 0.94 for sepsis diagnosis and 0.84 for mortality prediction. The combination of HSI and clinical data consistently outperformed clinical data alone, especially when only limited clinical data was available.
Functional Parameter Analysis
Septic patients and non-survivors exhibited significantly lower tissue oxygen saturation and higher tissue hemoglobin and water content compared to non-septic patients and survivors. These findings align with known sepsis pathophysiology, where tissue hypoxia and edema formation are key drivers of organ dysfunction.
Comparison with Clinical Biomarkers and Scores
The HSI + clinical data models outperformed widely used clinical biomarkers and scores, such as the quick Sequential Organ Failure Assessment (qSOFA) score, Systemic Inflammatory Response Syndrome (SIRS) criteria, and various inflammatory biomarkers like procalcitonin (PCT) and C-reactive protein (CRP).
Overall Conclusion
This study demonstrates the feasibility of using deep learning-based HSI analysis for automated, non-invasive, and rapid sepsis diagnosis and mortality prediction in ICU patients. The proposed HSI biomarkers showed high predictive performance, further enhanced by incorporating minimal clinical data. These biomarkers outperformed widely used clinical scores and biomarkers, highlighting their potential for various clinical settings, including resource-limited scenarios and time-critical situations. Future work should focus on external validation, exploring the use of HSI in different clinical environments, and investigating its application in pediatric populations and longitudinal studies.
References
- Sepsis-3 criteria
- Global sepsis mortality statistics
- Importance of early sepsis diagnosis
- Challenges in sepsis management
- Sepsis pathophysiology
- Microcirculatory monitoring in sepsis
- Machine learning models for sepsis prediction
- Limitations of EHR-based models
- Previous HSI studies in sepsis
- Deep learning architectures and training methodologies