Authors:
Arya Hadizadeh Moghaddam、Mohsen Nayebi Kerdabadi、Cuncong Zhong、Zijun Yao
Paper:
https://arxiv.org/abs/2408.09635
Introduction
Background
Lung cancer remains one of the leading causes of death worldwide. According to the CDC, in 2020, there were 47 new cases of lung cancer and 32 related deaths per 100,000 individuals in the United States. Early detection is crucial for improving survival rates, and DNA microarray technology has emerged as a powerful tool for this purpose. DNA microarrays can measure the activity of tens of thousands of genes simultaneously, providing valuable insights into the aberrant gene expression profiles of cancer cells.
Problem Statement
Despite the potential of DNA microarrays, the limited number of samples in gene expression datasets poses a significant challenge. This “small data” dilemma restricts the application of complex methodologies like deep neural networks, which require large datasets for effective training. Traditional models like support vector machines (SVMs) are often preferred due to their ability to handle smaller datasets, but they fall short in capturing the intricate interplay among high-dimensional gene expression signatures.
To address this challenge, this study introduces a meta-learning-based approach to enhance lung cancer detection from gene expression profiles. By leveraging multiple datasets, meta-learning aims to optimize machine learning models for better generalization and quicker adaptation to target datasets with limited samples.
Related Work
Gene Expression Profiling
Gene expression profiling using DNA microarrays has been extensively studied for cancer detection. Previous research has demonstrated the effectiveness of gene expression signatures in distinguishing between cancer patients and controls. However, the limited availability of samples in clinical studies remains a significant hurdle.
Transfer Learning
Transfer learning has been a popular approach to address the small data problem. It involves a two-stage learning pipeline where models are pre-trained on diverse datasets and then fine-tuned on the target dataset. While beneficial, transfer learning still requires abundant source datasets and considerable variability in downstream datasets, which may not always be available.
Meta-Learning
Meta-learning, or “learning to learn,” has emerged as a promising framework for rapid adaptation to target datasets using a small number of learning instances. Unlike transfer learning, meta-learning inherently optimizes for adaptability during the training phase, making it particularly effective for harnessing small-scale, high-dimensional gene expression data.
Research Methodology
Meta-Learning Framework
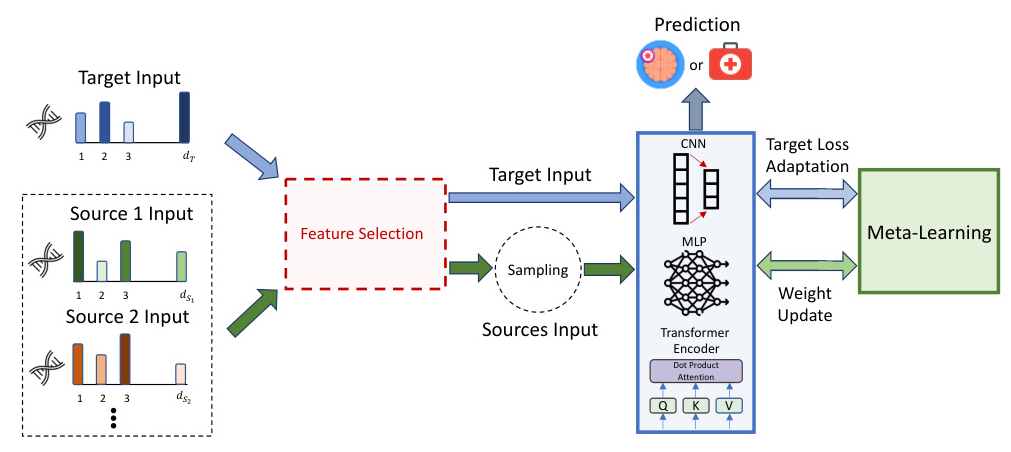
The proposed methodology employs a Model-Agnostic Meta-Learning (MAML) strategy to train sophisticated neural networks for lung cancer detection from gene expression samples. The framework involves the following steps:
- Data Collection: Four real-world gene expression datasets related to lung cancer and other conditions are used. Three datasets serve as sources, and one as the target for meta-learning.
- Feature Selection: A feature selection process identifies the most informative features while reducing dimensionality.
- Model Training: Neural networks receive samples from source data and target batches. Loss values from both datasets are used in meta-learning optimization.
- Prediction: The trained model is used for predicting outcomes in the target task.
Data Description
The datasets used in this study are:
- GSE132558: Samples from the University of Pennsylvania Medical Center and New York University Medical Center.
- GSE135304: Samples from multiple clinical sites, including the Helen F. Graham Cancer Center and New York University Langone Medical Center.
- GSE12771: Blood samples from smokers participating in epidemiological trials in Europe.
- GSE42830: Samples from patients diagnosed with various pulmonary diseases, including tuberculosis, sarcoidosis, pneumonia, and lung cancer.
Experimental Design
Planning and Designing the Experiment
The experiment involves using one dataset as the target and the others as sources in the context of meta-learning. The evaluation methodology employs 10-fold cross-validation, where 10% of the samples are reserved for testing, and the remaining 90% are used for training.
Data Preparation
The datasets are processed to ensure compatibility with the neural network architecture. This involves:
- Feature Selection: Identifying common genes across all datasets and leveraging the BioGrid database to focus on genes involved in genetic interactions.
- Normalization: Standard normalization is applied to enhance embedding learning.
Model Architectures
Three neural network architectures are employed:
- Multi-Layer Perceptron (MLP): Comprising multiple linear layers with LeakyReLU activation.
- Convolutional Neural Networks (CNNs): Incorporating convolutional and max-pooling layers for multi-level feature extraction.
- Transformer’s Encoder: Utilizing self-attention mechanisms to find interdependencies among input features.
Loss Function
The neural networks are optimized using a binary cross-entropy loss function. The meta-learning approach involves computing a meta-loss that combines the influences of both source and target datasets.
Results and Analysis
Performance Evaluation
The performance of the proposed meta-learning approach is evaluated against traditional statistical methods and deep learning models without meta-learning. The evaluation metrics include Accuracy, Precision, Recall, F1-score, and PRAUC (area under the precision-recall curve).
Comparative Analysis
The results demonstrate that the meta-learning approach consistently outperforms traditional methods and deep learning models without meta-learning. The Transformer’s encoder shows superior performance across all datasets.
Ablation Studies
Ablation studies reveal that meta-learning significantly enhances performance, especially on smaller datasets. The impact of varying the scaling factor (λ) in the meta-loss function is also analyzed, showing a trade-off between leveraging source data and incorporating target data.
Transfer Learning Comparison
The proposed meta-learning approach is compared with transfer learning. The results indicate that meta-learning exhibits greater generalization capabilities for cancer detection tasks based on gene expression levels.
Explainability
SHAP values are used to identify important features contributing to the decision-making process. The meta-learning approach changes the feature ranking and their respective impacts influenced by source datasets.
Overall Conclusion
This study presents a meta-learning framework for enhancing lung cancer detection from gene expression profiles. By leveraging multiple datasets, the proposed approach addresses the small data dilemma and improves the generalization of neural networks. The results demonstrate the superiority of meta-learning over traditional methods, deep learning techniques, and transfer learning. This research contributes to the development of more effective predictive models for cancer detection, ultimately aiding in early diagnosis and personalized treatment.
Acknowledgments
This work was funded in part by the National Science Foundation (NSF) under award number DBI-1943291, and by the University of Kansas New Faculty Research Development (NFRD) Award.