Authors:
Arthur Cerveira、Frederico Kremer、Darling de Andrade Lourenço、Ulisses B Corrêa
Paper:
https://arxiv.org/abs/2408.10482
Introduction
The field of drug discovery is undergoing a significant transformation with the advent of Artificial Intelligence (AI) techniques. These computational methods are increasingly being used to design and predict the properties of new therapeutic molecules. Traditional drug discovery methods often focus on single-target drugs, but this approach has limitations, especially for complex diseases like those affecting the central nervous system. Multi-target Drug Discovery (MTDD) aims to develop drugs that can modulate multiple targets simultaneously, offering potential advantages such as improved efficacy and reduced side effects. However, there is a lack of standardized benchmarks to evaluate the effectiveness of AI tools in designing multi-target drugs. This study proposes an evaluation framework for AI-driven molecular design in MTDD scenarios, using brain diseases as a case study.
Related Work
Multi-target Drugs for Complex Diseases
Multi-target drugs (MTDs) are emerging as a promising alternative for treating complex diseases such as central nervous system disorders, cancer, immune diseases, and cardiovascular diseases. Unlike target-specific drugs (TSDs), MTDs aim to modulate multiple targets simultaneously, addressing the complex nature of many physiological processes. However, designing effective MTDs is challenging due to the complexity of biological systems and the potential for off-target effects. Despite these challenges, well-designed MTDs offer significant advantages, including synergistic effects, enhanced therapeutic efficacy, and reduced likelihood of drug resistance.
Assessment of de Novo Molecular Design Strategies
De Novo Molecular Design (dNMD) techniques are evaluated using two main approaches: distribution-learning and goal-directed generation. Distribution-learning tasks assess the quality and diversity of generated molecules compared to the training set, while goal-directed generation focuses on optimizing molecules to meet a desired molecular profile. Popular benchmark suites like Guacamol and Molecular Sets (MOSES) are used to evaluate these techniques. However, there is currently no established goal-directed benchmark for assessing dNMD methods in MTDD tasks.
Artificial Intelligence Techniques for Molecular Design
AI-driven dNMD approaches, including Evolutionary Algorithms (EA) and Deep Generative Models (DGM), have shown promising results in designing early-stage therapeutic compounds. These strategies must generate and score new molecules, optimizing their exploration based on scoring outcomes. The scoring function may comprise multiple objectives and criteria that should be optimized simultaneously.
QSAR Models for Drug Discovery
Quantitative Structure-Activity Relationship (QSAR) modeling is an in silico technique used to estimate a molecule’s properties based on its structure. QSAR models are built using statistical or machine learning algorithms and training data from in vitro tested molecules. These models can predict the biological activity of a molecule against a disease protein target, aiding in the drug discovery process.
Research Methodology
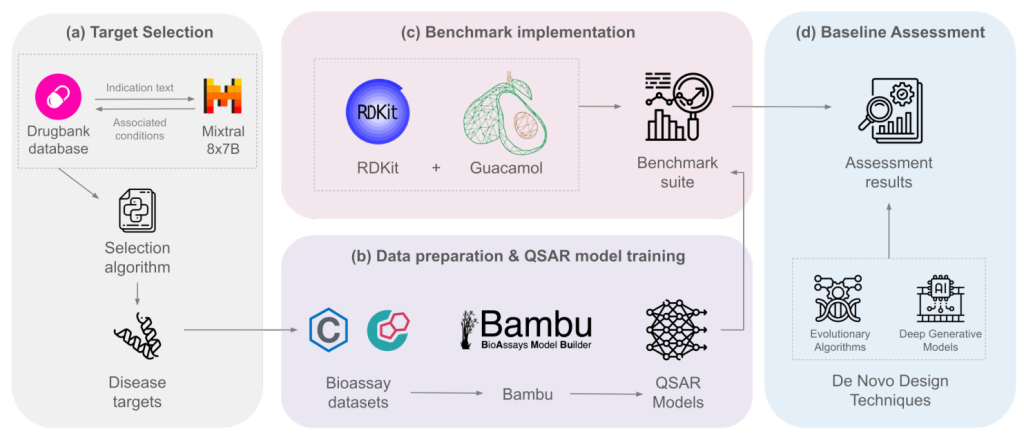
Target Selection
The target selection methodology employs the DrugBank database to identify the most common target combinations for each disease. An LLM is used to perform a structured extraction of associated diseases for each small molecule drug in the database. The co-occurrence matrix of protein targets is computed, and a greedy algorithm selects the best combination of targets. The Mixtral 8x7B LLM from Mistral AI is used for information extraction, and the targets are classified into Alzheimer’s, Schizophrenia, Parkinson’s, and Others.
Data Preparation
Training QSAR models requires quality datasets containing molecules and their measured in vitro activity. Bioassay datasets are collected from the PubChem platform and enriched with information from ChEMBL. A drug-like molecule database is derived from the ChEMBL 24 compound library, processed to remove salts, neutralize charges, and filter molecules based on specific criteria.
QSAR Model Training
QSAR models are trained using Bambu, an AutoML package for QSAR models. The Lo-Hi Splitter method is used for model evaluation and data split, simulating a real-world lead optimization scenario. Various machine learning algorithms and hyperparameter configurations are evaluated, and the best model is selected based on performance metrics.
Benchmark Implementation
QSAR models predict molecule activity against defined targets, and the scores are aggregated through a geometric mean operation. Physicochemical properties are evaluated using the CNS MPO and the ability to cross the Blood-Brain Barrier (BBB). The synthetic accessibility score (SAScore) is calculated to discriminate feasible molecules from infeasible ones. The final score estimates the molecule’s potential as an early-stage candidate for MTD against the specified disease.
Assessing the Baseline Techniques
Four dNMD models and algorithms are assessed: two LSTM models (LSTM-HC and LSTM-PPO) and two EA-based strategies (SMILES GA and Graph GA). The highest-scoring molecule from the processed dNMD dataset is used as a baseline for comparison. Hyperparameter setups are configured for a fair comparison, and the performance of each strategy is evaluated.
Experimental Design
Alzheimer’s Disease MPO Benchmark
Alzheimer’s Disease (AD) is a multifactorial neurodegenerative disease. The properties considered for the AD MPO benchmark include activity against pathological proteins, ability to cross the BBB, CNS MPO physicochemical properties, and synthetic accessibility score. The molecular targets are AChE and MAO-B.
Schizophrenia MPO Benchmark
Schizophrenia is a complex neuropsychiatric disorder. The properties considered for the schizophrenia MPO benchmark include molecule response against associated receptors, ability to cross the BBB, CNS MPO physicochemical properties, and synthetic accessibility score. The targets are dopamine D2 receptor (D2R) and serotonin 5-hydroxytryptamine 2A receptor (5-HT2AR).
Parkinson’s Disease MPO Benchmark
Parkinson’s Disease (PD) is characterized by the progressive degeneration of dopaminergic neurons. The MPO benchmark for PD includes evaluating a compound’s ability to modulate defined targets, penetrate the BBB, adhere to CNS MPO guidelines, and achieve a synthetic accessibility score. The targets are dopamine D2 receptors (D2R) and dopamine D3 receptors (D3R).
Results and Analysis
QSAR Models Evaluation
The QSAR models were trained using an AutoML workflow, and the best model was selected based on performance metrics. The selected models are tree-based ML algorithms, which capture nonlinear relationships between features and the target variable.
Disease-guided Benchmarks
The results for each dNMD method on the proposed benchmarks are displayed in Table III. The Graph GA technique outperformed other methods for AD and PD MPO benchmarks, while LSTM-HC achieved the best Schizophrenia MPO results. Both EAs and DGMs achieved competitive results on individual scoring functions, and most dNMD strategies outperformed the Best of Dataset in the final score.
Overall Conclusion
This study proposed a novel evaluation framework for AI-driven molecular design methods in MTDD scenarios. The benchmarking methodology assesses crucial aspects of the drug discovery process, including target modulation, physicochemical properties, and synthetic accessibility. The framework was implemented using brain diseases as a case study, and the results demonstrated the efficacy of both EA- and DGM-based approaches. The proposed framework can be extended to various diseases and relevant properties, providing a practical tool for assessing dNMD strategies and developing potential early-stage therapeutic compounds. The insights from this work can guide future strategies for designing dNMD techniques directed to MTDD use cases.