Authors:
Xin Wang、Xiaoyu Liu、Peng Huang、Pu Huang、Shu Hu、Hongtu Zhu
Paper:
https://arxiv.org/abs/2408.08881
Introduction
Background
Medical image segmentation is a critical task in clinical practice, aiding in precise medicine, therapeutic outcome assessment, and disease diagnosis. It involves delineating organ boundaries and pathological regions, which enhances anatomical understanding and abnormality detection. Traditional segmentation models often struggle with the complex patterns and minute variations in medical images, which are crucial for clinical diagnosis.
Problem Statement
Despite the advancements in foundation models like MedSAM, accurately assessing the uncertainty of their predictions remains a significant challenge. Understanding and managing this uncertainty is essential for improving the robustness and trustworthiness of these models in practical applications.
Related Work
Foundation Models for Medical Image Segmentation
Foundation models such as MedSAM have been developed to address the limitations of early segmentation models. These models are more robust, efficient, and applicable to various data modalities and situations. However, the uncertainty associated with their learning process on diverse datasets has not been thoroughly investigated.
Uncertainty-aware Learning
Incorporating uncertainty-aware learning into segmentation models can enhance their performance by balancing different aspects of the segmentation tasks (e.g., boundary, pixel, region). This approach can address issues like class imbalance and improve the overall robustness of the models.
Research Methodology
Proposed Model: U-MedSAM
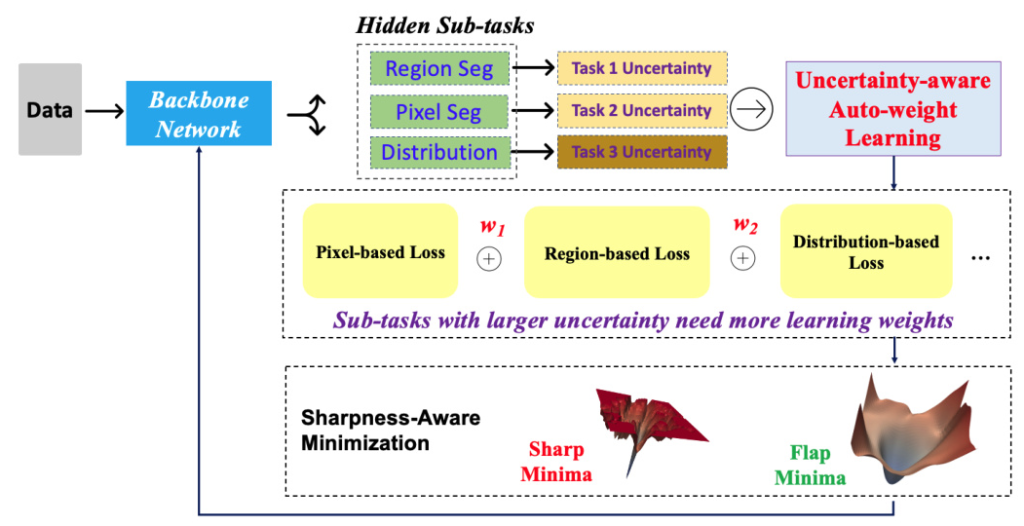
The U-MedSAM model extends the MedSAM architecture by integrating an uncertainty-aware loss function and the Sharpness-Aware Minimization (SharpMin) optimizer. This combination aims to enhance segmentation accuracy and robustness.
Uncertainty-aware Loss Function
The uncertainty-aware loss function in U-MedSAM combines three components:
1. Pixel-based Loss: Measures differences at the pixel level (e.g., Mean Squared Error loss).
2. Region-based Loss: Used for region segmentation (e.g., Dice coefficient loss).
3. Distribution-based Loss: Compares predicted and ground truth distributions (e.g., Cross-entropy loss and Shape-Distance loss).
Sharpness-Aware Minimization (SharpMin)
SharpMin improves generalization by finding flat minima in the loss landscape, thereby reducing overfitting. This optimizer guides the model towards parameter values that result in consistent performance across diverse data samples.
Experimental Design
Dataset and Evaluation Measures
The model was developed using the challenge dataset, with evaluation metrics including the Dice Similarity Coefficient (DSC) and Normalized Surface Dice (NSD). These metrics, along with running time, contribute to the overall ranking computation.
Implementation Details
The development environment included Ubuntu 20.04.1 LTS, Intel Xeon CPU, 125GB RAM, and four NVIDIA Titan Xp GPUs. The model was implemented using Python 3.10.14 and the PyTorch framework.
Training Protocols
The training protocols followed the LiteMedSAM implementation, with specific adjustments for the U-MedSAM model. The training involved 135 epochs with an initial learning rate of 0.00005, using the AdamW optimizer.
Results and Analysis
Quantitative Results
The results demonstrated that incorporating the uncertainty-aware loss and SharpMin optimization improved segmentation accuracy. Training with only the SD loss yielded an 83.86% DSC, while the uncertainty-aware loss increased DSC to 85.48%. Incorporating SharpMin further enhanced DSC to 86.10%.
Qualitative Results
Qualitative results showed that U-MedSAM produced clearer and more accurate segmentation boundaries compared to the baseline models. Examples of well-segmented images from various modalities highlighted the model’s effectiveness.
Segmentation Efficiency
The running speed of U-MedSAM was compared to the LiteMedSAM baseline, demonstrating competitive performance in terms of efficiency.
Limitations and Future Work
Despite achieving promising dice scores, the model’s speed was not optimized. Future work will explore model compression techniques such as quantization and tensor compression to enhance efficiency. Additionally, other advanced methods will be investigated to further improve performance and speed.
Overall Conclusion
U-MedSAM is an innovative model designed for robust medical image segmentation. By combining the MedSAM architecture with an uncertainty-aware learning framework and the SharpMin optimizer, U-MedSAM achieves superior accuracy and robustness. Comparative analysis with top-performing models highlights its effectiveness in enhancing segmentation performance.
Acknowledgements
The authors thank all data owners for making the medical images publicly available and CodaLab for hosting the challenge platform. Xin Wang is supported by the University at Albany Start-up Grant.