Authors:
Huy Quoc To、Ming Liu、Guangyan Huang
Paper:
https://arxiv.org/abs/2408.10729
Introduction
The rapid advancement of large language models (LLMs) has revolutionized the ability to process complex information across various fields, including science. The exponential growth of scientific literature, such as the 2.4 million scholarly papers on ArXiv and 36 million publications on PubMed, has enabled these models to effectively learn and understand scientific knowledge. However, the substantial computational resources, data, and training time required for LLMs pose significant challenges. This review aims to summarize the current advances in making LLMs more accessible for scientific applications and to explore cost-effective strategies for their deployment.
Related Surveys
Several surveys have explored pre-trained language models (PLMs) for science and methods to make LLMs more accessible. For instance, Ho et al. (2024) provided a comprehensive review of scientific language models (SciLM), evaluating their performance across various domains and tasks. Wang et al. (2023) focused on the biomedical sector, while Kalyan et al. (2022) explored transformer-based PLMs in the biomedical domain. Wan et al. (2024b) and Xu et al. (2024) discussed the resource-intensive nature of LLMs and proposed methods to improve their efficiency, such as knowledge distillation and model-centric approaches.
Advancement in Efficient LLMs for Science
Biology
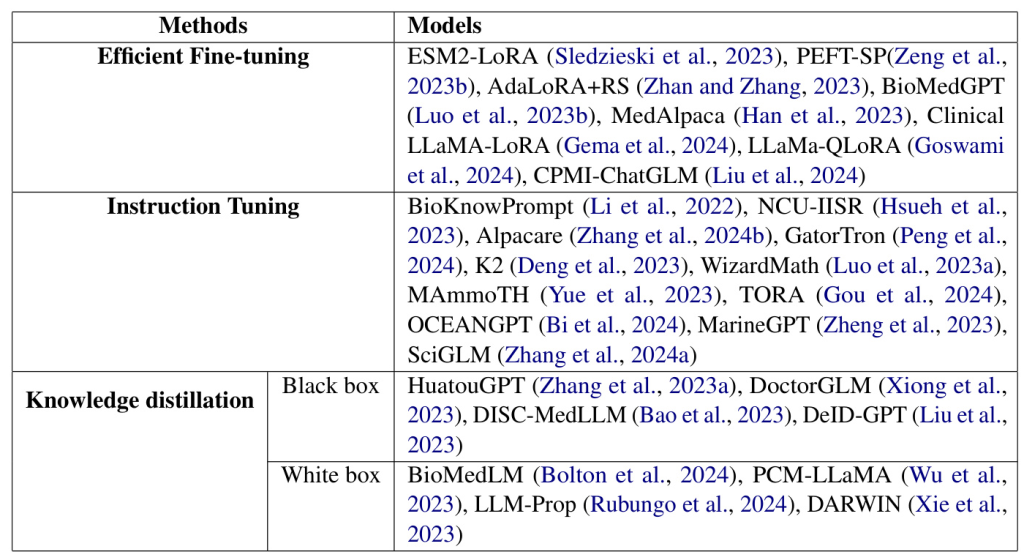
In biology, parameter-efficient fine-tuning (PEFT) techniques have been employed to address the computational challenges of large models. For example, Sledzieski et al. (2023) used PEFT on the ESM2 model to predict protein-protein interactions, achieving superior performance with less memory usage. Other studies, such as Zeng et al. (2023b) and Zhan and Zhang (2023), explored various PEFT techniques for tasks like signal peptide prediction and genomic language understanding.
Biomedical Domain
The biomedical field has seen significant advancements with LLMs like GPT-4 and Med-PaLM 2. However, these models are resource-intensive. Researchers have developed cost-effective solutions, such as BioKnowPrompt (Li et al., 2022) and HuatuoGPT (Zhang et al., 2023a), which leverage unlabelled data and data augmentation techniques. Other approaches, like DoctorGLM (Xiong et al., 2023) and BioMedLM (Bolton et al., 2024), focus on instruction-tuning and knowledge distillation to enhance performance while reducing resource requirements.
Clinical Domain
In the clinical domain, PEFT frameworks have been developed to adapt LLMs for clinical tasks. Gema et al. (2024) introduced a two-step PEFT framework for clinical outcome prediction, while Goswami et al. (2024) used prompt engineering and fine-tuning for summarizing hospital discharge summaries. Liu et al. (2024) created CPMI-ChatGLM for recommending Chinese patent medicine, demonstrating the potential of LLMs in traditional Chinese medicine research.
Mathematics
LLMs like GPT-4 have shown remarkable abilities in mathematical reasoning. Luo et al. (2023a) introduced WizardMath, which improves mathematical reasoning using Reinforcement Learning from Evol-Instruct Feedback (RLEIF). Yue et al. (2023) presented MAmmoTH, a series of open-source LLMs designed for mathematics, achieving significant accuracy improvements on mathematical benchmarks.
Geoscience
Deng et al. (2023) unveiled K2, the first LLM specifically designed for geoscience applications. They created GeoSignal and GeoBench datasets for geoscience instruction tuning and evaluation, demonstrating the effectiveness of their approach in enhancing geoscientific research.
Chemistry
In chemistry, researchers have focused on using textual descriptions of crystal structures for property prediction. Rubungo et al. (2024) introduced LLM-Prop, a method employing LLMs to predict crystal properties based on textual descriptions, outperforming domain-specific models like MatBERT.
Ocean Science
Ocean science has seen the introduction of OCEANGPT (Bi et al., 2024) and MarineGPT (Zheng et al., 2023), the first LLMs specifically designed for ocean science and marine domains. These models aim to enhance the understanding and application of oceanographic knowledge.
Multi-Scientific Domains
Xie et al. (2023) introduced DARWIN, a series of LLMs optimized for scientific disciplines like material science, chemistry, and physics. Zhang et al. (2024a) presented SciGLM, a suite of scientific language models designed for college-level scientific reasoning, addressing data scarcity challenges in the science domain.
Challenges and Future Directions
Data Collection
The lack of labeled data is a common issue in training language models for scientific fields. Current approaches like active learning and in-context learning have partially alleviated this problem, but they still rely heavily on human involvement. Future research should focus on developing methods to efficiently utilize unlabeled data.
Data Selection
Determining the optimal data volume and filtering out low-quality data are ongoing challenges. Recent methods like AlpaGasus and Superfiltering have shown promise, but further research is needed to establish clear guidelines for data selection in scientific domains.
Utilizing Multiple LLMs
Integrating knowledge from multiple LLMs into a single model can enhance performance. Techniques like knowledge fusion and multi-agent interaction graphs have shown potential, but extensive research is still needed in the scientific community.
Addressing Catastrophic Forgetting
Continual learning approaches like Lifelong-MoE and DCL aim to dynamically enhance models while preserving prior knowledge. However, preserving the initial model’s abilities and transferring knowledge across different domains remain significant challenges.
Multimodality
Integrating multi-modal information into scientific language models is essential for research progress. Developing efficient strategies to enhance the accessibility of LLMs and incorporate multimodality in scientific domains is crucial.
Further Reducing the Cost
Despite the impressive capabilities of modern LLMs, their substantial resource demands highlight the need for effective solutions. Methods like quantization, parameter pruning, and memory-efficient fine-tuning offer viable solutions, but further research is needed to make LLMs more accessible and sustainable.
Conclusion
The rapid progress of large language models has greatly improved our ability to tackle intricate tasks in scientific research. However, the substantial resource requirements of these models pose significant challenges. This review highlighted various cost-effective techniques for utilizing LLMs in scientific domains and addressed the challenges in fully harnessing their potential. Ensuring broader accessibility and applicability of LLMs in scientific research remains a critical goal.
Limitations
This review primarily focuses on text-based scientific information, setting aside other forms such as images, videos, audio, and structured knowledge like knowledge graphs and databases. The review highlights advancements from 2023 and 2024, but may have missed some recent studies. Future work should consider these limitations and explore additional methodologies.