Authors:
Stefano Woerner、Christian F. Baumgartner
Paper:
https://arxiv.org/abs/2408.08058
Introduction
Machine learning has significantly advanced medical imaging and diagnostics, but these advancements often rely on large, well-annotated datasets. For many medical applications, especially rare diseases, collecting such datasets is challenging. Few-shot learning (FSL) offers a potential solution by enabling the training of models with minimal data. This study benchmarks the performance of various foundation models in FSL and zero-shot learning (ZSL) across diverse medical imaging datasets.
Methods
Dataset
The study utilizes the MedIMeta dataset, which includes 19 publicly available datasets covering 10 different imaging modalities. This standardized meta-dataset allows for a comprehensive evaluation of FSL and ZSL performance across various medical imaging tasks.
Simulation of FSL and ZSL Tasks
FSL tasks are simulated by randomly sampling labeled training samples and unlabeled query samples from each dataset. The study evaluates performance across different numbers of labeled samples (n ∈ {1, 2, 3, 5, 7, 10, 15, 20, 25, 30}). For ZSL, task instances with n = 0 are used.
Pretrained Models
The study evaluates three pretraining paradigms: supervised pretraining, self-supervised pretraining, and contrastive language-image pretraining (CLIP). The models include:
- Fully Supervised Models: ResNet (18, 50, 101) and Vision Transformer (ViT) with variations pretrained on ImageNet and ImageNet21k.
- Self-supervised Models: DINOv2 models (ViT-B, ViT-L, ViT-g) pretrained using self-supervised knowledge distillation.
- Contrastive Language-Image Pretraining: CLIP models (ViT-B, ViT-L, ViT-H, ViT-g) pretrained on large datasets like LAION-2B, and BiomedCLIP pretrained on medical data.
Few-shot Learning Strategies
Two adaptation strategies are evaluated: fine-tuning and linear probing. Fine-tuning involves training all weights in the network, while linear probing trains only a new classification layer on the final representations of the network.
Zero-shot Learning Strategy
For ZSL, CLIP and BiomedCLIP models use text prompts to classify images without labeled training examples. Three prompt templates are tested to evaluate ZSL performance.
Metrics
Performance is measured using the area under the receiver operator curve (AUROC) averaged over 100 task instances for each dataset and training set size. The harmonic mean of the AUROCs from each dataset provides an average performance measure.
Experiments and Results
Hyperparameter Optimization
The optimal hyperparameters were similar across models, with Adam as the best-performing optimizer and zero initialization for the classification head. A learning rate of 10^-4 with at least 120 training steps was generally optimal.
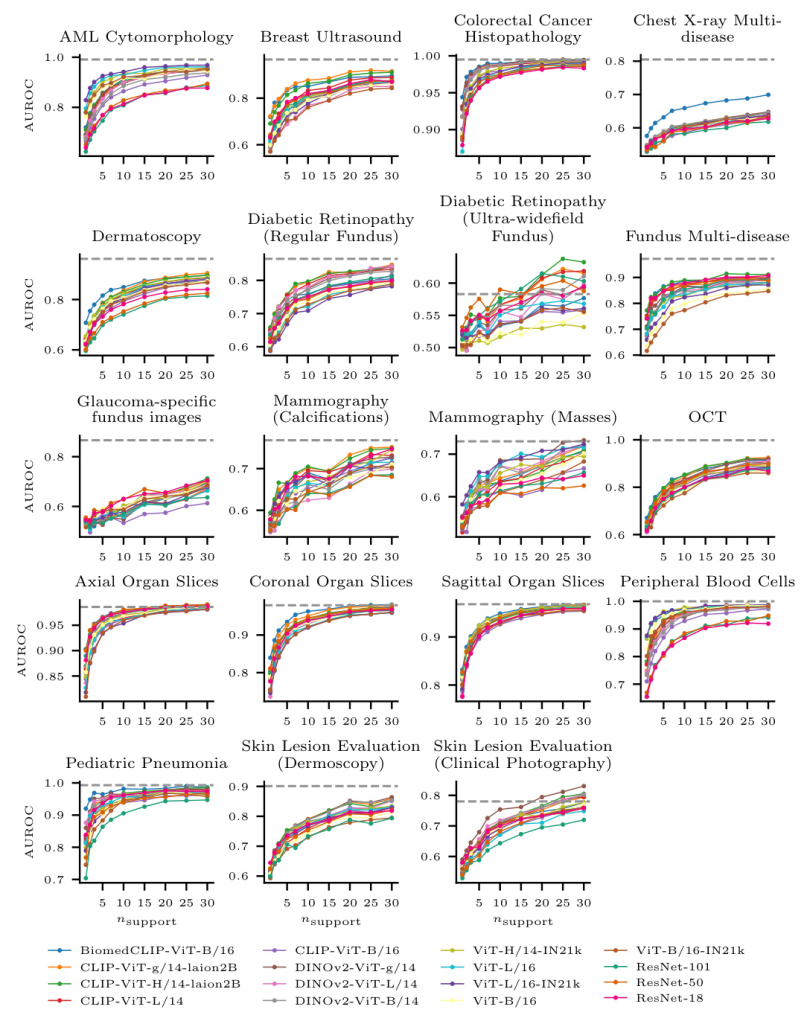
Few-shot Learning Performance
- Linear Probing: BiomedCLIP and CLIP-ViT-H performed best on average. CLIP-ViT-H outperformed other models for n ≥ 7, while BiomedCLIP excelled for smaller n.
- Dataset-specific Performance: Performance varied across datasets. BiomedCLIP performed well on some datasets but poorly on others, likely due to the representation of images in the pretraining dataset.
- Comparison with Fully Supervised Learning: For some tasks, 30-shot FSL performance nearly matched fully supervised performance, suggesting that linear probing of foundation models can be a viable alternative to training from scratch with small datasets.
- Fine-tuning vs. Linear Probing: Linear probing on large models outperformed fine-tuning of smaller models. However, fine-tuning ResNet-18 performed well with more training data, indicating that lower network complexity may be preferable in FSL scenarios.
Zero-shot Learning Performance
ZSL performance was significantly lower than FSL performance, indicating that ZSL may not yet be suitable for general medical image analysis tasks.
Model Complexity and Pretraining Data Size
There was a strong positive correlation between model size, pretraining set size, and few-shot performance. BiomedCLIP, despite having fewer parameters and a smaller pretraining dataset, performed well, highlighting the importance of domain-specific pretraining.
Conclusion
This study provides a comprehensive benchmark of FSL and ZSL performance across diverse medical imaging datasets. Key findings include:
- BiomedCLIP is the best strategy for very low data regimes (n ≤ 5).
- CLIP-ViT-H performs better with more data.
- Fine-tuning ResNet-18 is a strong alternative given sufficient data.
- Large foundation models trained on non-medical data perform well on medical tasks, but domain-specific models like BiomedCLIP show significant potential.
Further research is needed to develop foundation models tailored for medical applications and to collect more diverse medical datasets.