Authors:
Paper:
https://arxiv.org/abs/2408.06618
Generalized Knowledge-Enhanced Framework for Biomedical Entity and Relation Extraction
Introduction
The rapid increase in biomedical publications has made it challenging for researchers to stay updated with the latest findings. To address this, various frameworks have been developed for automatic extraction of biomedical entities and their relations. This paper introduces a novel framework that leverages external knowledge to construct a reusable background knowledge graph for biomedical entity and relation extraction. The framework is inspired by how humans learn domain-specific topics, starting with foundational knowledge and extending to specialized topics.
Related Works
Advanced strategies for domain-specific document understanding involve learning from both input texts and external domain knowledge. Several knowledge-enhanced models have been developed based on this approach:
- KnowBertAttention: Utilizes SciBERT for token-level representations and introduces external knowledge from UMLS.
- KECI: A knowledge-enhanced model with collective inference that integrates multi-relational graph structures and global information.
KECI Model Framework
- Initial Processing: Uses text embeddings to construct a knowledge graph with feature vectors for biomedical entities.
- External Knowledge Integration: Establishes a background knowledge graph using external sources like UMLS.
- Feature Integration: Combines feature vectors from the task dataset with those from the external knowledge dataset for final predictions.
Motivations and Contributions
Existing approaches often waste external knowledge and resources due to task-dependent loss functions. This paper introduces a knowledge-enhanced model that builds a general knowledge graph (GK) reusable across different tasks. The framework separates general knowledge (GK) from specific knowledge (SK), optimizing knowledge utilization and enhancing scalability.
Key Contributions
- Task-Independence and Reusability: Develops a GK component with reusable feature vectors applicable across different biomedical tasks.
- Efficient Knowledge Utilization: Separates GK and SK components to minimize redundancy and enhance scalability.
- Focused and Adaptive Modeling: Fuses GK with SK components for effective learning across different biomedical domains.
- Competitive Benchmarking: Demonstrates competitive performance on BioRelEx and ADE datasets.
Methods
Overview
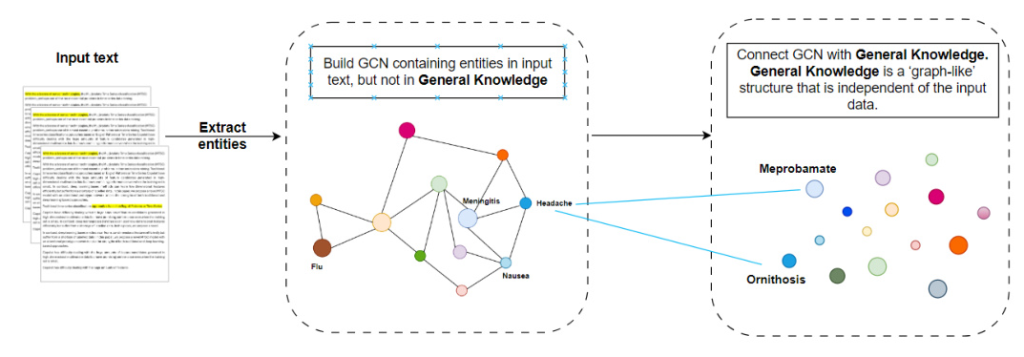
The framework builds a generalized background knowledge graph with two components: General-Knowledge (GK) and Specific-Knowledge (SK). The GK component encapsulates reusable domain knowledge, while the SK component is tailored to individual tasks.
General-Knowledge (GK) Component
The GK component is constructed by extracting relational data using BioBERT and training a feed-forward neural network (FFNN) to encode relational weights into entity representations.
Extracting Relational Data
BioBERT is used to extract relational information from entities within the knowledge source data. Masking techniques form hypothetical sentences to infer relational information, and geometric vector operations calculate the expected weight indicating how likely a relation matches a subject-object pair.
Entities Representation Training
Based on the extracted weights, an FFNN is trained to encode relational weights into entity representations. The final embedding for each entity is a concatenation of the initial embedding from BioBERT and the relational embedding obtained from training.
Specific-Knowledge (SK) Component and Fusion with GK Component
The SK component is built using a graph neural network (GCN) on entities from input documents for a specific task. An additional GCN connects nodes in the specific task’s graph to those in the GK component.
Experiments
Data Sources and Tasks
Two data sources are used for the GK component: UMLS and Wikidata. The framework is evaluated using two biomedical datasets: BioRelEx and ADE.
Baselines
The framework is compared against several state-of-the-art methods, including Relation-Metric, SpERT, SPANMulti-Head, SentContextOnly, FlatAttention, KnowBertAttention, and KECI Full Model.
Entities and Relations Extraction Results
The framework demonstrates competitive performance on both ADE and BioRelEx datasets, outperforming several baselines.
Discussion
Relation Weights Extraction
The framework effectively predicts relation weights and provides related information, demonstrating its ability to capture relational information accurately.
Contribution of GK Component
The GK component significantly contributes to the overall knowledge graph, especially for frequent entities within the dataset. This highlights the framework’s ability to reuse knowledge across tasks.
Conclusion and Future Works
The paper introduces a knowledge-enhanced model with GK and SK components, demonstrating competitive results on ADE and BioRelEx datasets. Future work includes expanding the GK component and applying the framework to other research areas beyond biomedical fields.
The implementation is available at GitHub.