Authors:
Kun Luo、Bowen Zheng、Shidong Lv、Jie Tao、Qiang Wei
Paper:
https://arxiv.org/abs/2408.09746
Introduction
Prostate cancer is the second most common cancer and the fifth leading cause of cancer-related death among men worldwide. Early diagnosis is crucial for effective treatment and reducing mortality. Multiparametric magnetic resonance imaging (mpMRI) has become a popular non-invasive method for diagnosing prostate cancer, providing both anatomical and functional information. However, interpreting mpMRI requires significant expertise, highlighting the need for automated grading systems to assist radiologists.
The study titled “Enhanced Cascade Prostate Cancer Classifier in mp-MRI Utilizing Recall Feedback Adaptive Loss and Prior Knowledge-Based Feature Extraction” addresses the challenges of automated grading in mpMRI. The authors propose a solution that incorporates prior knowledge, addresses the issue of uneven medical sample distribution, and maintains high interpretability in mpMRI.
Related Work
Recent advancements in computer-aided diagnosis (CAD) for prostate cancer using mpMRI images have shown promising results. Conventional machine learning methods primarily use image-based approaches or radiomics for binary classification tasks. However, these methods often struggle with more complex multi-classification problems. Deep learning techniques have been extensively explored to address this issue, particularly in the context of multi-classification tasks.
Several studies have integrated prior knowledge into deep learning models to improve performance. For example, anatomical segmentation masks, prior diagnosis comparisons, and histopathology information have been used to guide the network’s focus and improve classification accuracy. However, these methods often use simplistic and superficial information, lacking the integration of complex diagnostic criteria and medical judgment.
The imbalanced sample sizes across diseases and stages can bias the classification towards specific samples. Prioritizing recall rates for these specific samples is crucial to avoid misdiagnosis. Various loss functions have been proposed to address this issue, such as the Recall Loss, which incorporates recall as a dynamic parameter. However, these methods may not fully suit prostate cancer ISUP classification, especially in distinguishing fewer samples.
Research Methodology
The authors propose a novel approach for prostate cancer ISUP classification by combining prior knowledge of grading criteria and diagnostic procedures. The key contributions of the study are as follows:
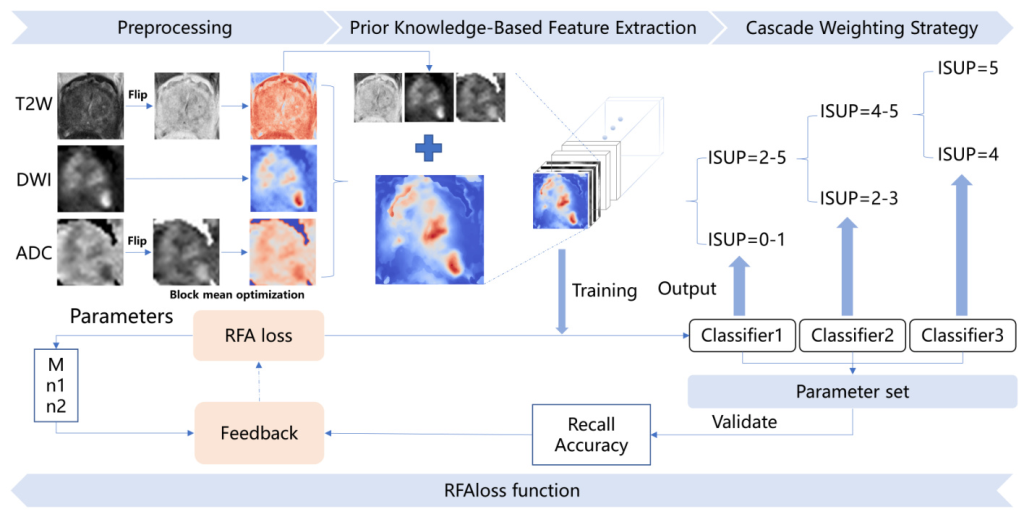
- Recall Feedback Adaptive Loss (RFAloss): A novel loss function that dynamically adjusts during training to expand the parameter search space and adjust the search direction based on recall feedback. This addresses the bias caused by imbalanced sample distribution.
- Prior Knowledge-Based Feature Extraction (F-E): A feature extraction strategy based on the Prostate Imaging Reporting and Data System (PI-RADS) criteria for prostate cancer in mpMRI. This strategy improves the model’s generalization ability and interpretability.
- Cascade Confidence Refinement Strategy: A strategy that allows the classifier to output and fuse results based on clinical practice, resulting in a more balanced confusion matrix even with highly imbalanced samples.
Experimental Design
Patient Data and Preprocessing
The study uses the public dataset of the PI-CAI Challenge, which consists of three sequences: T2WI, ADC, and DWI, along with prostate gland segmentation masks and their ISUP labels for 1499 cases. The data preprocessing steps include:
- Prostate Gland Cropping: Cropping and resampling the data to ensure that the effective training data are within the largest bounding cuboid of the prostate gland.
- ADC, T2W Image Signal Flipping: Performing signal flipping on the ADC and T2WI sequences to better model image features.
- Block Mean Optimization for Ineffective Region in ADC Images: Introducing a 2×2 block mean operator to mitigate the impact of extreme signal levels in ADC and DWI images.
Prior Knowledge-Based Feature Extraction (F-E)
The feature extraction strategy incorporates prior knowledge from the PI-RADS criteria. The key components of the F-E strategy are:
- Symmetrical Difference Algorithm: Quantifying the intensity differences in symmetrical positions to highlight signals in the feature area while suppressing signals in non-feature areas.
- Symmetrically Weighted Algorithm: Capturing the differences between the axial line and surrounding positions using a weighted sum and normalization approach.
- Feature Fusion Strategy: Fusing the results from the SW and SD algorithms using a normal distribution function to create the final feature map.
Recall Feedback Adaptive Loss (RFAloss)
The RFAloss function incorporates accuracy and recall as dynamic feedback to guide the search direction and expand the search scope. The key components of the RFAloss function are:
- Setting of Base Framework: Using the cross-entropy loss function as the base structure.
- Setting of Dynamic Differential Feedback Coefficients: Introducing dynamic differential feedback coefficients to guide the loss function towards the recall of the class of interest.
- Setting of Feedback Intensity Hyperparameters: Introducing three adjustable hyperparameters (M, n1, and n2) to control the feedback process.
Cascade Refinement Confidence Strategy
The cascade refinement strategy transforms the overall ISUP classification into a cascade classification task. Three classifiers are trained to perform binary classifications:
- Classifier1: Distinguishes between levels 0-1 and 2-5 for diagnosing csPCa.
- Classifier2: Separates levels 2-3 from levels 4-5 to determine appropriate clinical interventions.
- Classifier3: Focuses on classifying level 4 versus level 5 to quantify the severity of high-grade csPCa.
The cascade strategy refines the output probabilities of positive classes for each classifier, resulting in a balanced confusion matrix even with highly imbalanced samples.
Results and Analysis
Hyperparameter Experiments
The hyperparameter experiments evaluate the general effects of various hyperparameters of the RFAloss function. The results show that the performance of experiments with n1 and n2 initially improves but then declines as the variables change progressively. The recall, ARS, and F2-score achieve the best results when M is set to 0.3.
Ablation Experiments
The ablation experiments compare the proposed RFAloss function with classical loss functions such as cross-entropy loss, focal loss, and recall loss. The results demonstrate that the RFAloss function significantly improves the recall rate while achieving excellent accuracy. Applying both feature extraction and RFAloss simultaneously improves the F2-score, AUC, and recall rate compared to applying RFAloss alone.
Comparison Experiments
The comparison experiments evaluate the proposed method against three other methods: M3T, HiFuse, and MedViT. The results show that the proposed method significantly improves the recall rate while maintaining high accuracy and AUC.
Evaluation of Cascaded Refinement Confidence Strategy
The cascaded refinement strategy is evaluated using optimal parameters from ISUP 2-3 and 4-5 classification. The results show that the strategy renders the overall results more balanced and focuses more on the diagonal positions in the confusion matrix. The recall rate for each category decreases as the ISUP grade increases, but even for the most challenging category five, there is still a 40% recall rate.
Overall Conclusion
The study proposes a recall-guided deep learning-assisted ISUP grading strategy based on mpMRI. The key contributions include the Recall Feedback Adaptive Loss (RFAloss), Prior Knowledge-Based Feature Extraction (F-E), and Cascade Confidence Refinement Strategy. The proposed method significantly improves the recall rate while maintaining high accuracy and interpretability. The results demonstrate the practical significance of integrating ISUP grading indicators and diagnostic processes of prostate cancer into deep learning. The methods proposed in this study provide valuable references for medical image processing and its practical applications.