Authors:
Yang Nan、Huichi Zhou、Xiaodan Xing、Guang Yang
Paper:
https://arxiv.org/abs/2408.08704
Beyond the Hype: A Dispassionate Look at Vision-Language Models in Medical Scenarios
Introduction
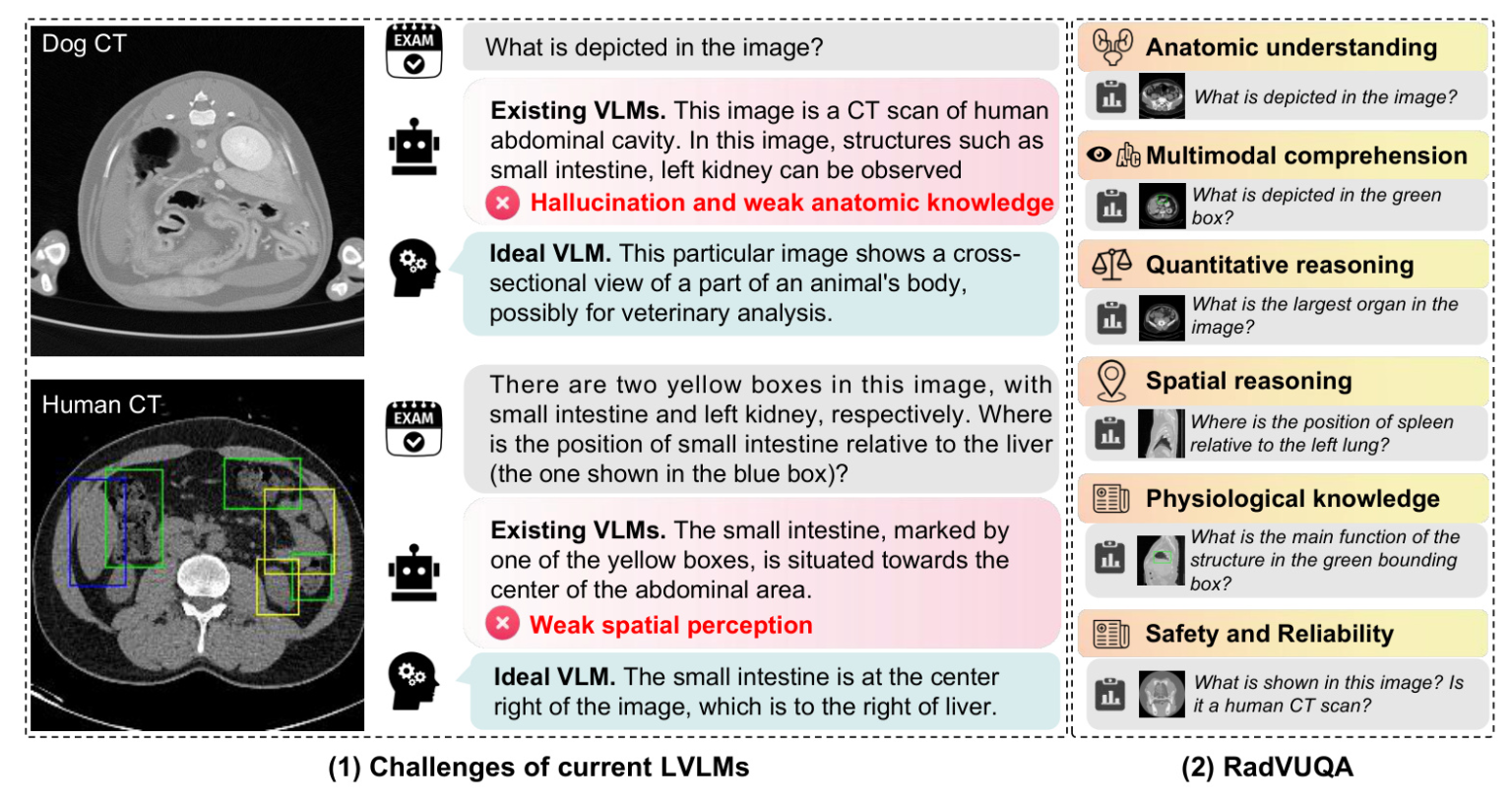
Recent advancements in Large Vision-Language Models (LVLMs) have showcased their impressive capabilities across various tasks. However, their performance and reliability in specialized domains such as medicine remain underexplored. This study introduces RadVUQA, a novel Radiological Visual Understanding and Question Answering benchmark, to comprehensively evaluate existing LVLMs. RadVUQA assesses LVLMs across five dimensions: anatomical understanding, multimodal comprehension, quantitative and spatial reasoning, physiological knowledge, and robustness. The findings reveal significant deficiencies in both generalized and medical-specific LVLMs, highlighting the need for more robust and intelligent models.
Related Works
Large Vision Language Models
Vision-language models have gained significant attention due to their ability to integrate visual and linguistic instructions. Models like CLIP, Flamingo, and BLIP2 have demonstrated impressive performance by mapping images and text into a shared embedding space. In the medical domain, models such as Med-Flamingo, LLaVA-Med, and RadFM have been developed, leveraging paired medical image-text data to enhance their capabilities. Despite these advancements, a comprehensive evaluation of generalized LVLMs in the medical domain remains lacking.
Medical VQA Benchmarks
Several benchmarks have been established to evaluate the capabilities of medical LVLMs, including VQA-RAD, Path-VQA, SLAKE, OmniMedVQA, and CAREs. These benchmarks have focused on various aspects such as anatomical understanding, abnormalities, and social characteristics. However, they often overlook critical capabilities like quantitative reasoning and multimodal comprehension. This study aims to address these gaps by providing a more comprehensive evaluation framework.
Methods
Data Resources and Data Preprocessing
RadVUQA was developed using multi-source, multi-anatomical public datasets, resulting in subsets RadVUQA-CT, RadVUQA-MRI, and RadVUQA-OOD. The dataset includes diverse representations of body parts and varied prompts, ensuring a comprehensive evaluation framework. Data preprocessing involved assigning labels to each instance based on spatial category, category, anatomical location, and general category, enabling accurate and semantically rich QA pairs.
Question Design
RadVUQA comprises open-ended and close-ended questions across various aspects:
- Anatomical Understanding: Evaluates the model’s ability to identify organs and structures.
- Multimodal Comprehension: Assesses the capability to interpret linguistic and visual instructions.
- Quantitative and Spatial Reasoning: Tests the model’s quantitative analysis and spatial perception.
- Physiological Knowledge: Investigates the model’s understanding of the functions and mechanisms of organs.
- Robustness: Simulates various scenarios to assess the model’s performance against unharmonised and synthetic data.
Experimental Settings and Metrics
Models were evaluated on two NVIDIA H100 NVL GPUs. The evaluation metrics included Response Accuracy (RA), Hallucination Score (HS), and Multiple-Choice Accuracy (MCA). Commercial large language models were used as evaluators to minimize subjective biases and reduce time costs.
Results
RadVUQA-CT
The performance of different LVLMs on CT data revealed that commercial models like GPT-4o and Gemini-Flash achieved the highest scores in various aspects. However, significant hallucination issues were observed, particularly in anatomic understanding and quantitative reasoning.
RadVUQA-MRI
Similar trends were observed in MRI data, with GPT-4o achieving state-of-the-art performance in most evaluation aspects. However, hallucination rates varied, with some models exhibiting more hallucinations in MRI data compared to CT data.
RadVUQA-OOD
The capabilities of LVLMs on unharmonised data showed that GPT-4o achieved the highest RA scores in open-ended questions, while Gemini-Flash obtained the best MCA scores in close-ended questions. However, hallucination issues persisted across different scenarios.
Discussion
Overall Performance
The overall performance of LVLMs remains stable across CT and MRI datasets, with commercial models outperforming open-source counterparts. However, significant gaps in foundational medical knowledge and robustness to unharmonised data were observed.
Sensitivity to CT Windowing
CT windowing significantly improved the performance of certain LVLMs in anatomic understanding and spatial reasoning, while others experienced negative effects. This suggests the need for more specific training resources and harmonisation of input data.
Effectiveness of CoT in Medical Q&A
Prompt-based Chain-of-Thought (CoT) strategies improved the performance of most LVLMs in open-ended questions, reducing hallucination frequencies and increasing accuracy. However, some models experienced increased hallucinations, indicating potential misjudgments.
Robustness to Unharmonised Data
LVLMs demonstrated varying tolerances to unharmonised data, with some models showing reduced accuracy but lower hallucination rates. This highlights the need for harmonisation foundation models to handle diverse clinical settings effectively.
Distinguishing Synthetic Data
LVLMs struggled to distinguish synthetic data from real ones, with low synthetic-detection rates observed across models. Integrating a refusal-to-answer strategy may enhance model safety and robustness.
Conclusion
This study presents RadVUQA, a comprehensive benchmark for evaluating LVLMs in medical scenarios. The findings reveal significant deficiencies in existing models, highlighting the need for more robust and intelligent LVLMs. Key insights include the potential of large-scale medical LVLMs, the benefits of clinical-specific techniques like CT windowing, and the effectiveness of Prompt-CoT strategies. However, challenges remain in handling unharmonised data and distinguishing synthetic data, underscoring the need for further research and development.