Authors:
Abdur R. Fayjie、Jutika Borah、Florencia Carbone、Jan Tack、Patrick Vandewalle
Paper:
https://arxiv.org/abs/2408.08432
Predictive Uncertainty Estimation in Deep Learning for Lung Carcinoma Classification in Digital Pathology under Real Dataset Shifts
Introduction
Deep learning (DL) has revolutionized various fields, including digital pathology and medical image classification. However, the reliability of DL models in clinical decision-making is often compromised due to distributional shifts in real-world data. This paper investigates the role of predictive uncertainty estimation in enhancing the robustness of DL models for lung carcinoma classification under different dataset shifts.
Background
Related Work
Uncertainty estimation in DL is crucial for providing calibrated predictions and enhancing model robustness. Bayesian Neural Networks (BNNs) are prominent in this domain but are computationally expensive. Alternative methods like Monte Carlo dropout (MC-dropout), deep ensembles, and few-shot learning (FSL) have been explored for uncertainty estimation in medical imaging.
Problem Set-up
The primary task is to classify lung adenocarcinoma in histopathology and radiology images. The study evaluates the performance and uncertainty of DL models under various distribution shifts, including in-distribution shifts and out-of-distribution (OOD) shifts.
Materials and Methodology
Uncertainty Estimation Methods
- MC Dropout: This method involves enabling dropout layers during test time to estimate uncertainty through multiple stochastic forward passes.
- Deep Ensemble: Multiple neural networks are trained independently, and their predictions are aggregated to estimate uncertainty.
- Few-Shot Learning (FSL): Prototypical Networks are used to classify images with limited labeled samples, leveraging previously acquired knowledge.
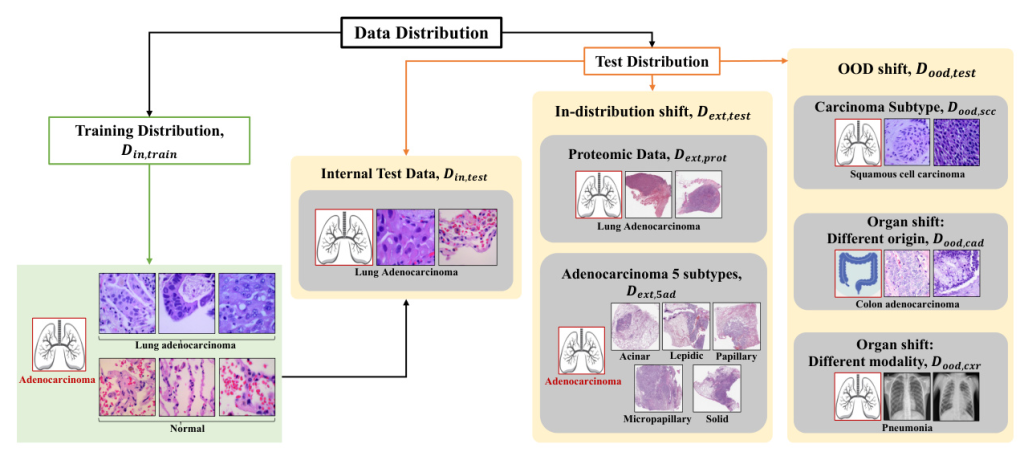
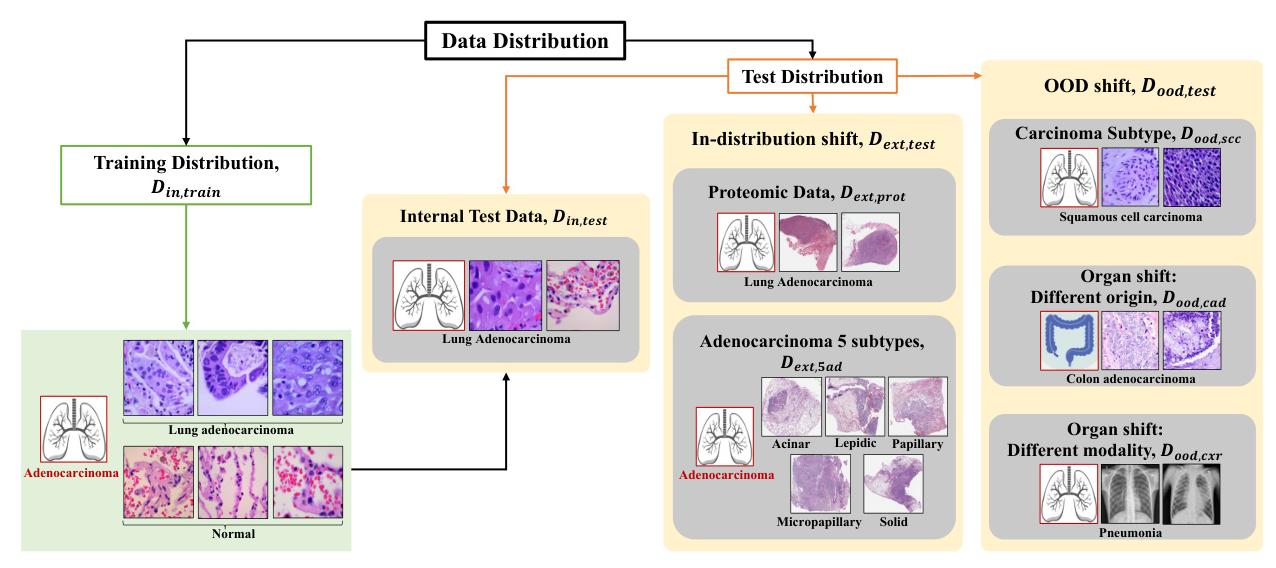
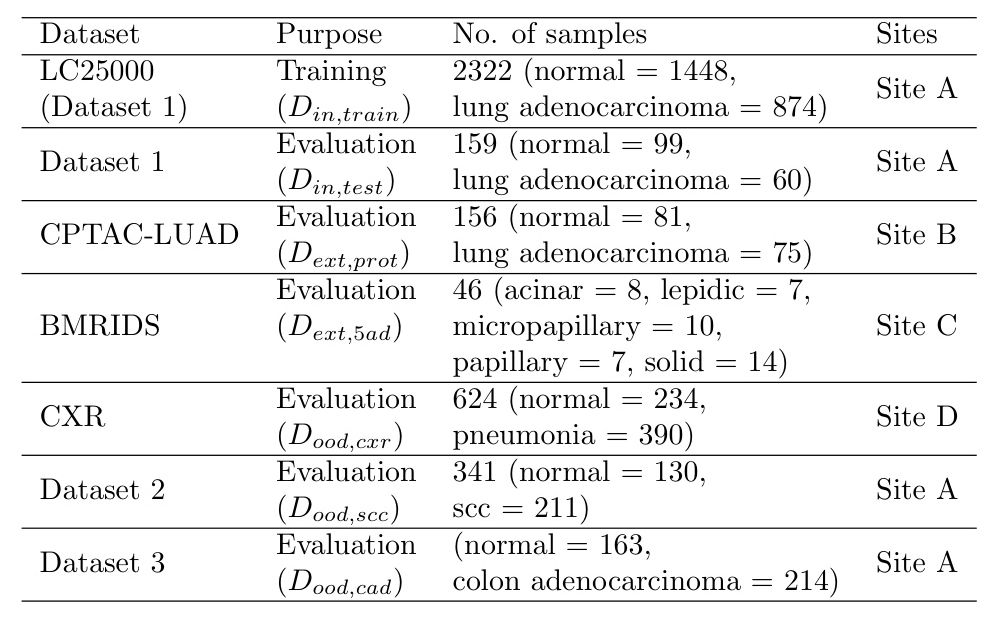
Datasets
The study uses several datasets, including LC25000, CPTAC-LUAD, BMIRDS, and Pneumonia Chest X-rays, to evaluate the models under different distribution shifts.
Uncertainty Metrics
Entropy is used as the primary metric for quantifying predictive uncertainty.
Evaluation Metrics
The models are evaluated using accuracy, area-under-the-receiver-operating-characteristic-curve (AUROC), and area-under-precision-recall (AUPR).
Experiments
Data Distribution Shifts
- In-distribution Shift (Dext,test): Variations within the same disease class, including different sub-types and proteomic data.
- Out-of-Distribution Shift (Dood,test): Completely different distributions, including different carcinoma sub-types, organ origins, and imaging modalities.
Implementation Details
- MC Dropout: ResNet50 model with dropout layers enabled during test time.
- Deep Ensemble: Five different neural networks (VGG19, ResNet50, DenseNet121, Xception, EfficientNetB0) trained independently.
- Few-Shot Learning: ResNet10 model trained using an episodic training strategy.
Experimental Results and Analysis
Evaluation on Predictive Performance under Dataset Shift
The models’ performance was evaluated on six different distributions. The results indicate a significant drop in performance when the data distribution shifts, especially for OOD shifts.
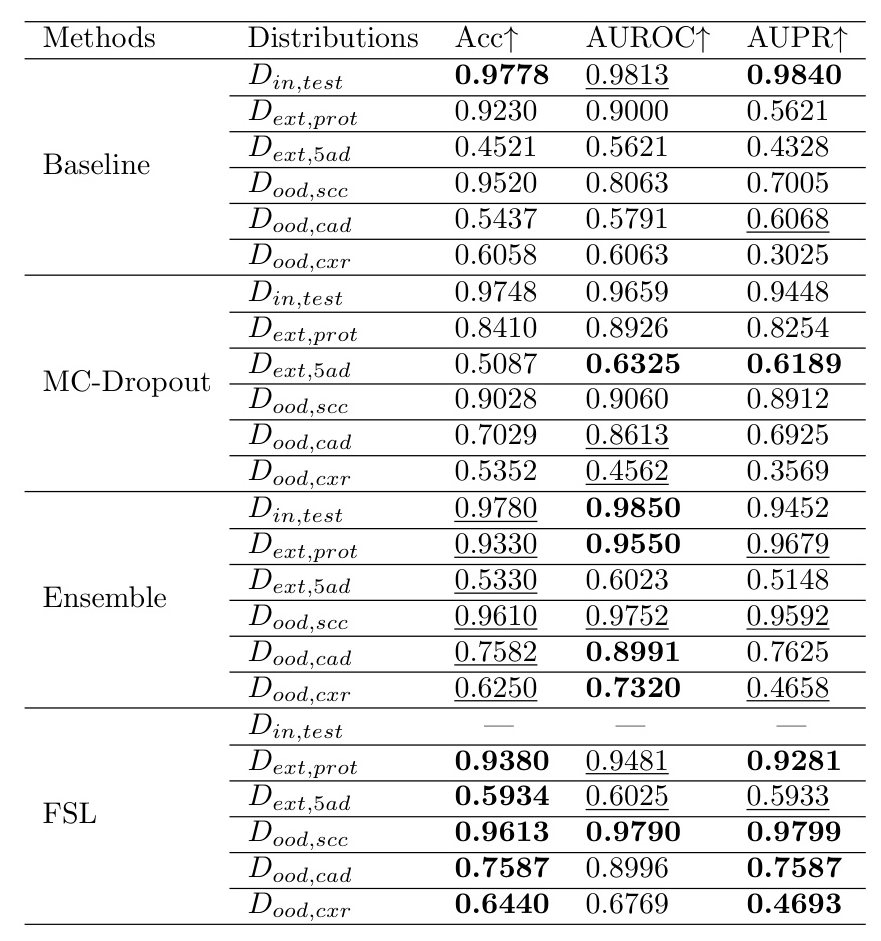
Evaluation on Predictive Uncertainty under Distribution Shifts
The entropy values were used to evaluate predictive uncertainty. The results show that the models exhibit higher uncertainty for OOD data, as expected.
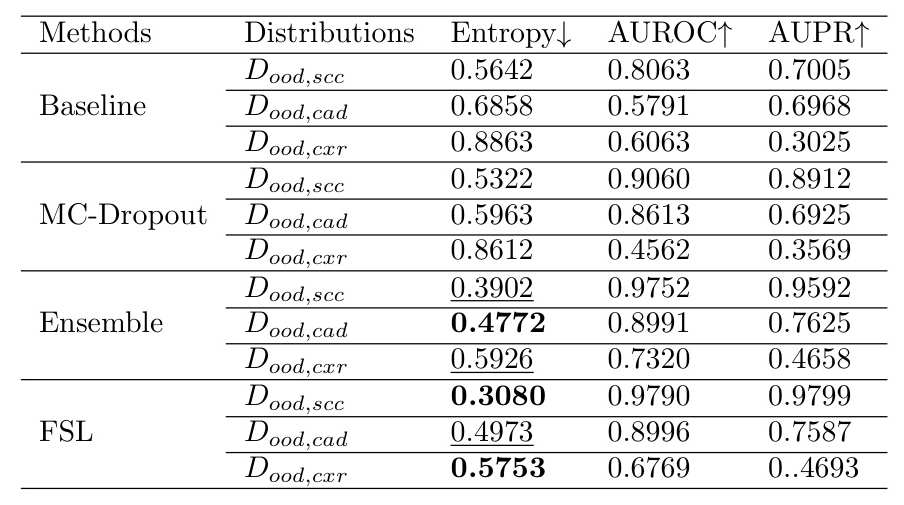
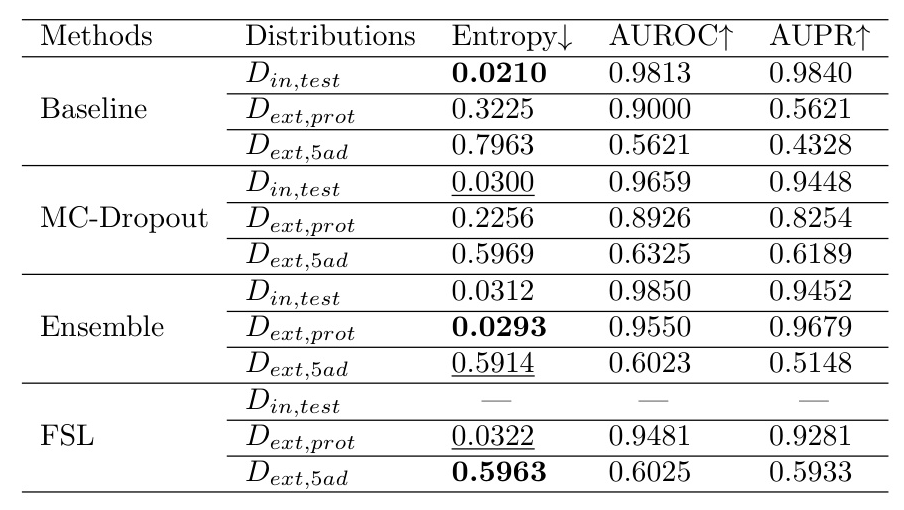
Evaluation on Different Carcinoma Sub-types
The models’ performance was also evaluated on different carcinoma sub-types. The results indicate that deep ensemble and FSL methods perform better in handling data shifts.
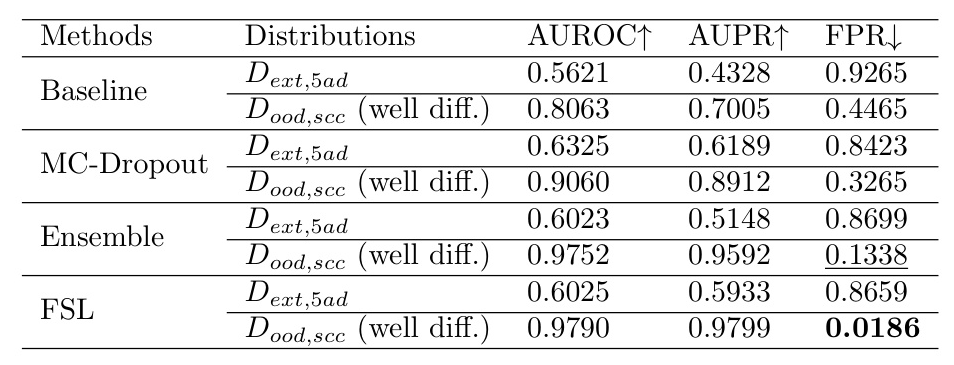
Discussion
The study demonstrates that predictive uncertainty estimation methods like deep ensemble and FSL can enhance the robustness of DL models in digital pathology. However, these methods also have limitations, such as higher computational costs for deep ensembles.
Future Prospects and Challenges
Future research should focus on integrating predictive uncertainty estimation into clinical decision-making systems. This includes addressing the diverse needs of healthcare and establishing trust between clinicians and DL models. Additionally, multi-institutional data representing patient diversity will be crucial for developing robust models.
Conclusion
This study highlights the importance of predictive uncertainty estimation in DL models for lung carcinoma classification. While the models perform well on in-domain data, their performance degrades under distribution shifts. Reliable uncertainty estimates can help integrate DL-based methods into clinical practices, provided they are used under the guidance of clinicians.
This blog post provides a detailed overview of the paper “Predictive Uncertainty Estimation in Deep Learning for Lung Carcinoma Classification in Digital Pathology under Real Dataset Shifts.” The study emphasizes the importance of uncertainty estimation in enhancing the robustness of DL models in clinical applications.