Authors:
Honggen Zhang、Xiangrui Gao、June Zhang、Lipeng Lai
Paper:
https://arxiv.org/abs/2408.09048
Introduction
Messenger RNA (mRNA)-based vaccines and therapeutics have revolutionized the pharmaceutical industry, offering new avenues for treating diseases such as COVID-19 and cancer. However, selecting optimal mRNA sequences from extensive libraries remains a costly and complex task. Effective mRNA therapeutics require sequences with optimized expression levels and stability. This paper introduces mRNA2vec, a novel contextual language model-based embedding method designed to enhance mRNA sequence representation by combining the 5’ untranslated region (UTR) and coding sequence (CDS) regions.
Related Work
mRNA Sequence-based Translation Prediction
The optimization of mRNA sequences to enhance expression and stability has become increasingly viable due to the availability of vast amounts of measured and synthesized sequences. Data-driven models have emerged as effective tools for predicting synonymous sequence expression, detecting highly nonlinear correlations between sequences and their functional outcomes. Notable works include:
- PERSIST-seq by Leppek et al. (2022), which predicts mRNA stability and ribosome load.
- Evolutionary algorithms by Diez et al. (2022) for codon optimization.
- Machine learning approaches by Cao et al. (2021) and Nieuwkoop et al. (2023) for modeling the 5’ UTR and CDS regions.
mRNA Sequence Representation Learning
Self-supervised learning and language models have recently emerged as powerful tools for extracting sequence features from large, unlabeled datasets. Key contributions include:
- DNA2vec by Ng (2017), inspired by word2vec, for learning distributed representations of k-mers.
- RNA-BERT by Akiyama and Sakakibara (2022), which adapts the BERT algorithm to non-coding RNA (ncRNA).
- CodonBERT by Li et al. (2023), which embeds codons to measure mRNA expression, stability, and immunogenicity.
Research Methodology
mRNA2vec Model
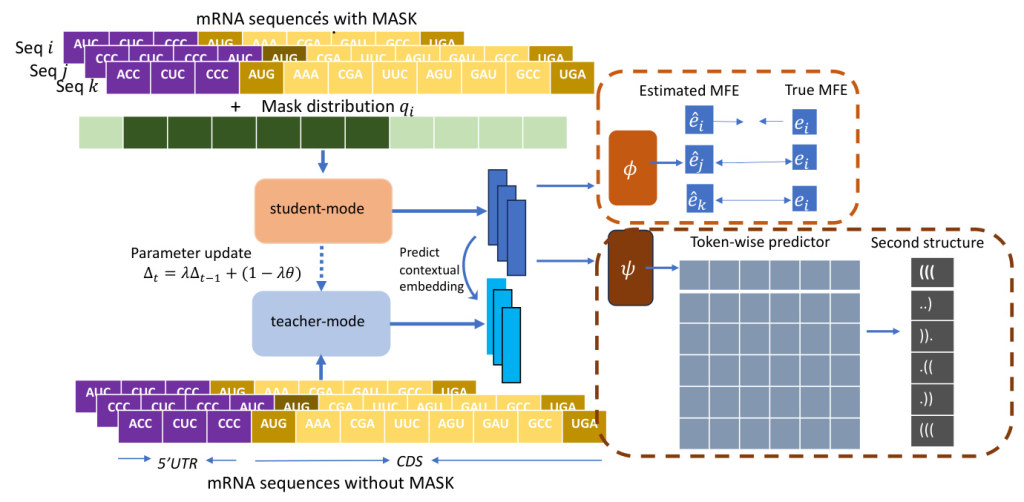
mRNA2vec is a novel mRNA language model that concatenates the 5’ UTR and CDS sequences to form a single input sequence. Unlike traditional models such as BERT or T5, which use masked sequences as input, mRNA2vec adopts a contextual target-based pre-training model inspired by data2vec. This approach allows the model to access both unmasked and masked sequences simultaneously.
Probabilistic Hard Masking Strategy
A probabilistic hard masking strategy is employed, where specific regions of the sequence, particularly biologically important areas, have a higher likelihood of being masked. This ensures that these critical regions are more effectively learned during training.
Auxiliary Pretext Tasks
Domain-specific information, such as Minimum Free Energy (MFE) and Secondary Structure (SS), is incorporated through the definition of auxiliary pre-training tasks. This integration strengthens the model’s ability to capture a representation across the entire sequence, enhancing its utility for translation-related downstream tasks on both 5’UTR and CDS regions.
Experimental Design
Dataset
For pre-training, mRNA sequences from five species (human, rat, mouse, chicken, and zebrafish) were collected from the NIH. The sequences were processed to extract the 5’UTR and CDS regions, resulting in 510k sequences with an average length of 459 bp. MFE values and SS were obtained using RNAfold from ViennaRNA.
For downstream tasks, three human cell types were used for the 5’ UTR data: Human embryonic kidney (HEK) 293T cell line, human prostate cancer cell line PC3 (PC3), and Human muscle tissue (Muscle). For the CDS data, the mRNA stability dataset and the mRFP Expression dataset were used.
Experiment Setting
The mRNA2vec pre-training architecture is built upon the data2vec framework, utilizing the T5 encoder as the student model. The model consists of 4 self-attention heads and 4 hidden layers, with each token represented by a 256-dimensional vector. The regressor and classifier are three-layer neural networks. AdamW was used as the optimizer, with a learning rate of 0.001 and a weight decay of 0.001. The maximum sequence length was set to 128, and the batch size was 256.
Results and Analysis
Pre-training Results
Different pre-training strategies were evaluated on mRNA sequences. The data2vec approach using T5 as the encoder showed better performance after several epochs on the 5’ UTR data compared to a standalone T5 encoder and an untrained model. Incorporating both MFE and SS losses consistently improved the base model across all downstream tasks.
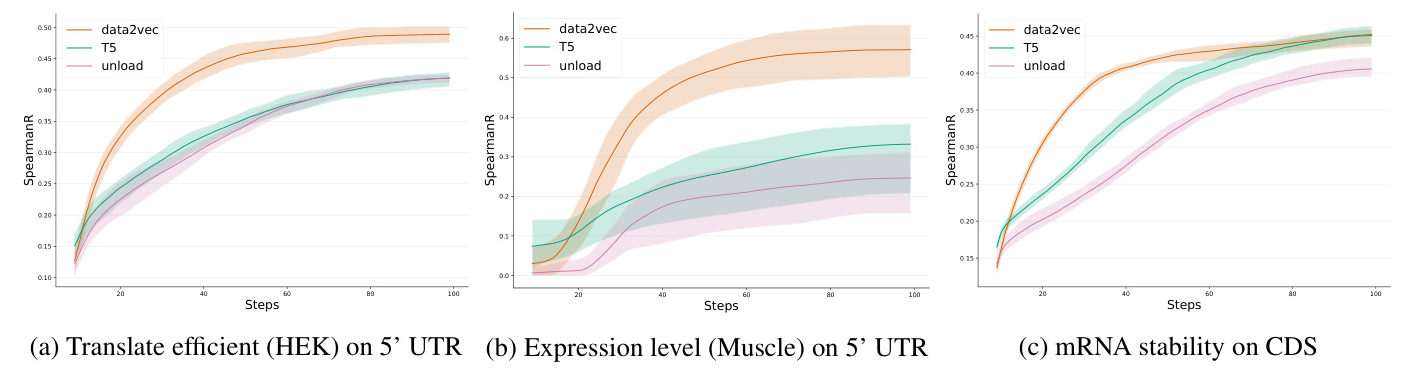
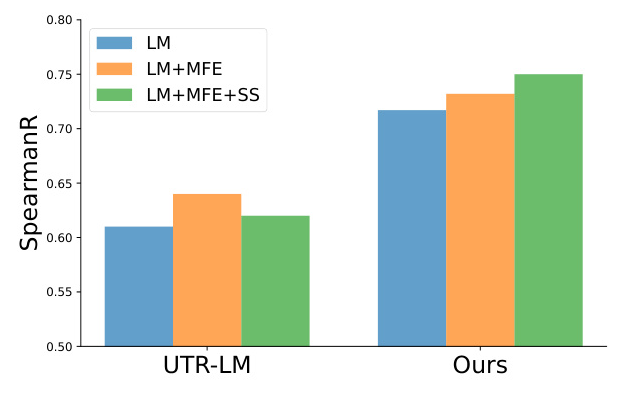
Downstream Tasks
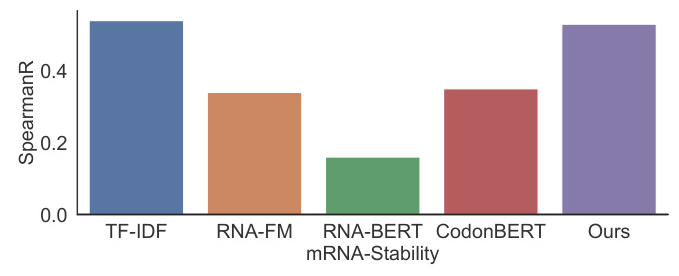
For the 5’UTR dataset, mRNA2vec outperformed all other approaches on datasets related to the Translation Efficiency (TE) and Expression Level (EL) tasks. For the CDS dataset, mRNA2vec significantly improved results compared to the codon representation method CodonBERT for mRNA stability and achieved comparable results for protein production levels.
Discussion
Key observations contributing to the final performance on downstream tasks include:
- Optimal Input Sequence Length: The input sequence length affects the TE outcome, with full sequences worsening the results. Selecting sub-sequences by truncating the input sequence or directly selecting the representation of the sub-sequence can improve performance.
- Selecting the Hidden State: Different hidden layers provide different representations of the mRNA sequence. Using the second last hidden state as the mRNA embedding improved performance.
- Designing a Better Downstream Task Regressor: More complex regressors generally improve performance over linear regressors, especially with larger datasets.
Overall Conclusion
mRNA2vec, a novel mRNA pre-training method based on the data2vec architecture and mRNA domain knowledge, demonstrates significant improvements in translation efficiency and expression level prediction tasks. The contextual learning and hard-mask strategy, along with the incorporation of domain-specific information such as MFE and SS, enhance the model’s ability to represent mRNA sequences effectively. mRNA2vec outperforms state-of-the-art benchmarks on TE and EL tasks and achieves comparable results on mRNA stability and protein production levels, showcasing its potential for advancing mRNA-based therapeutics and vaccine design.