Authors:
Tianyi Liu、Zhaorui Tan、Muyin Chen、Xi Yang、Haochuan Jiang、Kaizhu Huang
Paper:
https://arxiv.org/abs/2408.09465
Introduction
Brain tumors pose significant risks to human health, necessitating precise medical segmentation for effective treatment planning. Brain tumor segmentation typically relies on multiple magnetic resonance imaging (MRI) modalities, such as Fluid Attenuation Inversion Recovery (Flair), contrast-enhanced T1-weighted (T1ce), T1-weighted (T1), and T2-weighted (T2). These modalities complement each other, providing a comprehensive understanding of the tumor’s physical structure and physiopathology. However, in clinical practice, certain MRI modalities may be missing due to data corruption or variations in scanning protocols, presenting a challenge for accurate segmentation.
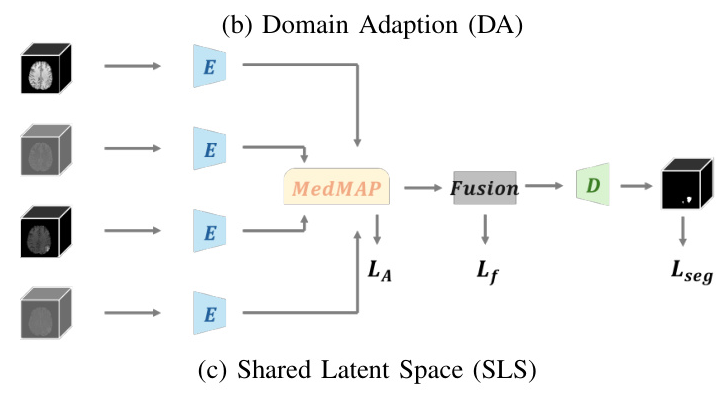
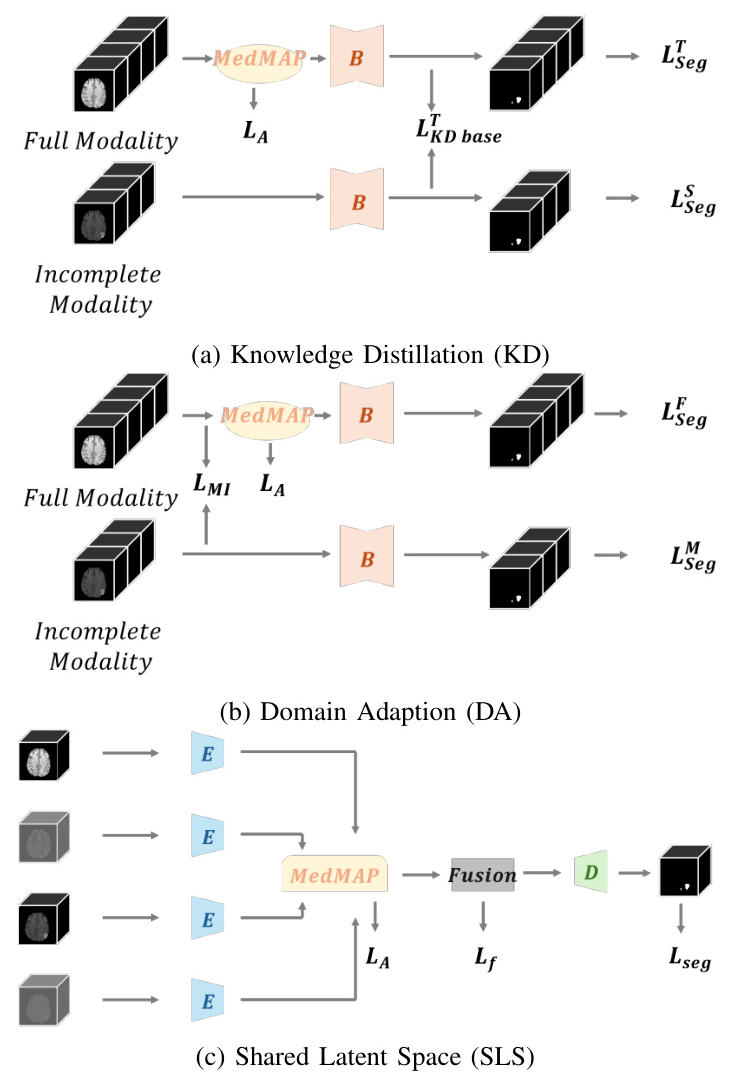
To address this issue, strategies like Knowledge Distillation (KD), Domain Adaptation (DA), and Shared Latent Space (SLS) have been employed. However, these methods often overlook the gaps between modalities, leading to suboptimal performance. This paper introduces a novel paradigm, the Medical Modality Alignment Paradigm (MedMAP), which aligns latent features of different modalities to a well-defined distribution anchor, thereby minimizing modality gaps and improving segmentation performance.
Related Work
Multi-modal Learning for Missing Modalities
Brain tumor segmentation methods often utilize multiple MRI modalities to achieve better performance. However, missing modalities can significantly degrade segmentation accuracy. Previous approaches to handle missing modalities can be categorized into three main strategies:
- Knowledge Distillation (KD): KD-based methods transfer knowledge from a teacher model trained on complete modalities to a student model trained on incomplete modalities. These methods often fail to capture feature invariance across modalities.
- Shared Latent Space (SLS): SLS methods aim to create a common representation space shared among all modalities. However, they often do not sufficiently eliminate modality gaps.
- Domain Adaptation (DA): DA methods minimize the gap between models trained on complete and incomplete modalities. These methods also often overlook the gaps between different modalities within complete modality models.
Alignment in Multi-domain Generalization
Domain generalization techniques aim to reduce gaps in the latent space across different data domains. These methods learn domain-independent representations to improve generalization. Inspired by these techniques, MedMAP aims to align latent distributions in the teacher model to reduce modality gaps and improve segmentation performance in missing modality scenarios.
Research Methodology
Theoretical Motivation
The primary goal of MedMAP is to minimize modality gaps in the latent space while maintaining prediction performance. The proposed alignment paradigm involves aligning each modality’s latent features to a pre-defined distribution, Pmix. This approach ensures a tighter Evidence Lower Bound (ELBO), theoretically certifying its effectiveness.
Aligning Medical Multi-modalities
MedMAP aligns the latent features of different modalities to a shared space using a feature encoding pipeline. The alignment is achieved by minimizing the Kullback-Leibler (KL) divergence between the latent features and the pre-defined distribution Pmix. Two forms of Pmix are proposed: Pk_mix (a specific modality’s latent distribution) and P*_mix (a weighted mixture of all modalities’ latent distributions).
Experimental Design
Datasets
The BraTS2018 and BraTS2020 datasets, consisting of 285 and 369 subjects respectively, are used for evaluation. Each subject includes four MRI modalities (T1, T1ce, T2, and Flair) and ground truth segmentation labels for different tumor sub-regions.
Implementation
The proposed alignment paradigm is integrated into several state-of-the-art backbones (PMKL, mmFormer, and ACN) for comparison. The models are implemented using PyTorch and trained on an Nvidia GeForce RTX 3090Ti GPU. The performance is evaluated using the Dice Score, with higher scores indicating better segmentation accuracy.
Results and Analysis
Quantitative Comparisons
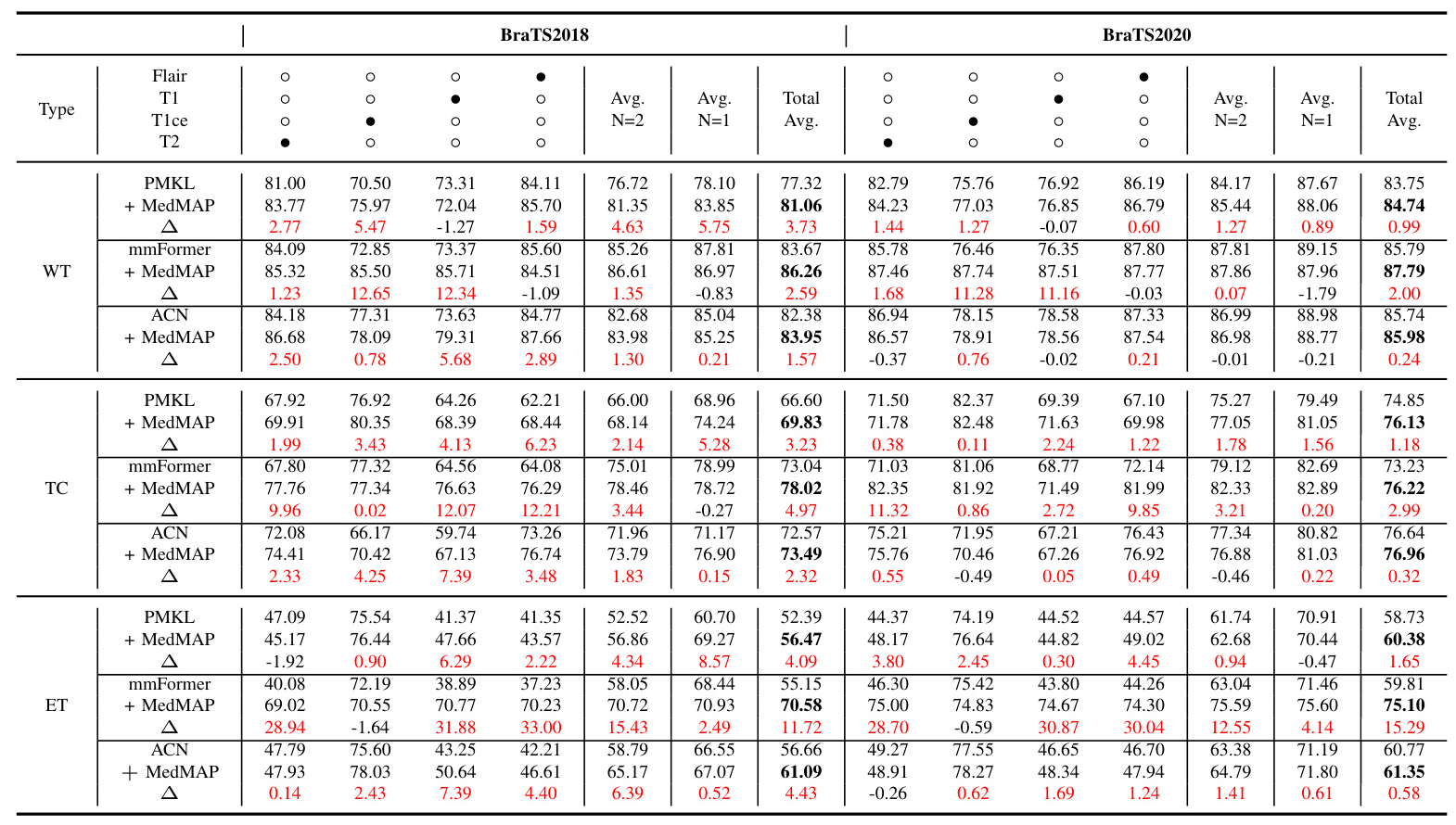
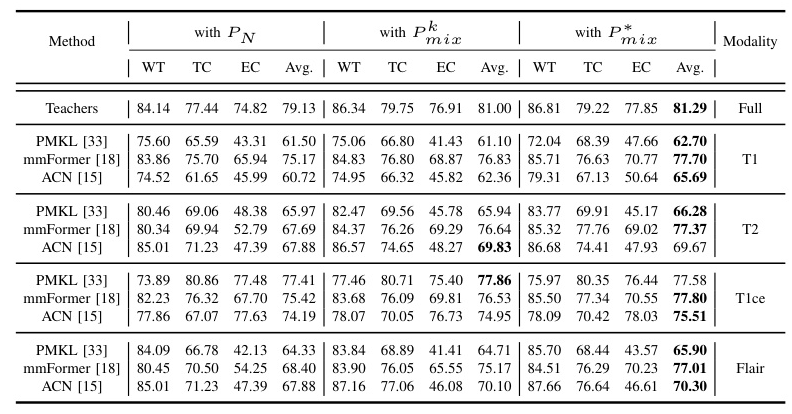
The proposed MedMAP significantly improves segmentation performance across different backbones and datasets. For instance, on the BraTS2018 dataset, MedMAP improves the Dice Scores by 3.68%, 7.67%, and 2.30% for PMKL, mmFormer, and ACN, respectively. Similar improvements are observed on the BraTS2020 dataset.
Performance Improvement on Different Prediction Classes
MedMAP enhances the prediction performance for different tumor sub-regions (WT, TC, and ET) across various missing modality scenarios. The improvements are particularly notable for challenging cases with multiple missing modalities.
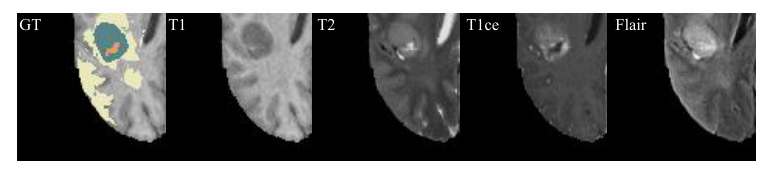
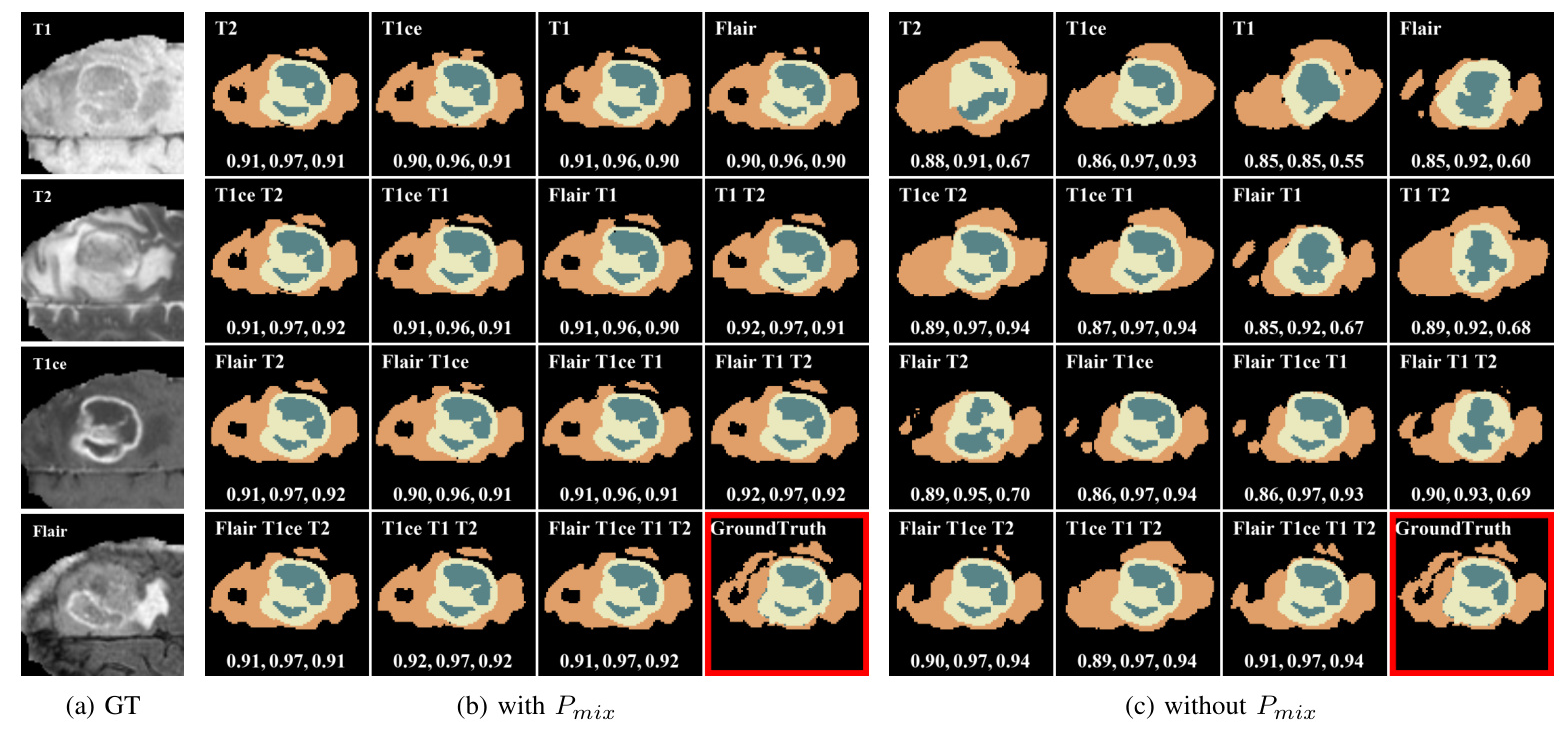
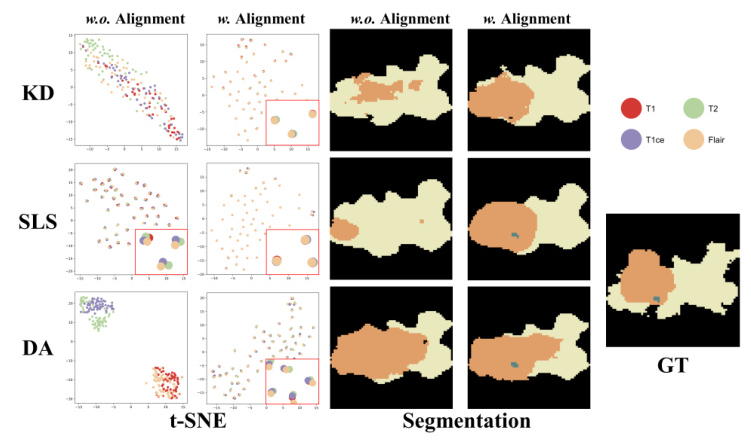
Qualitative Comparison
Visual comparisons demonstrate that models with MedMAP produce more accurate and detailed segmentation results, especially in cases with multiple missing modalities.
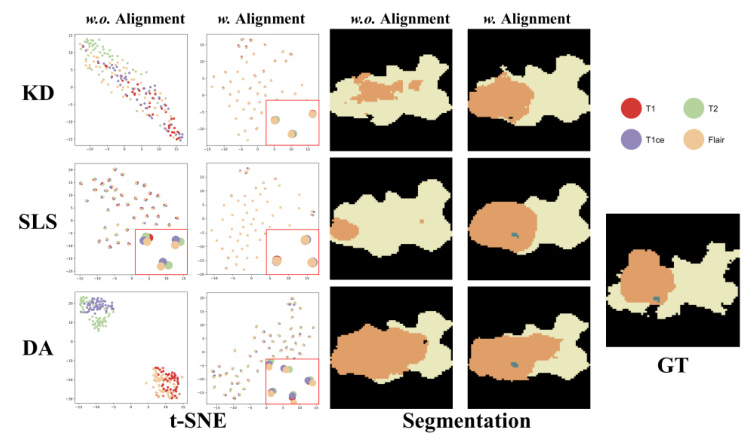
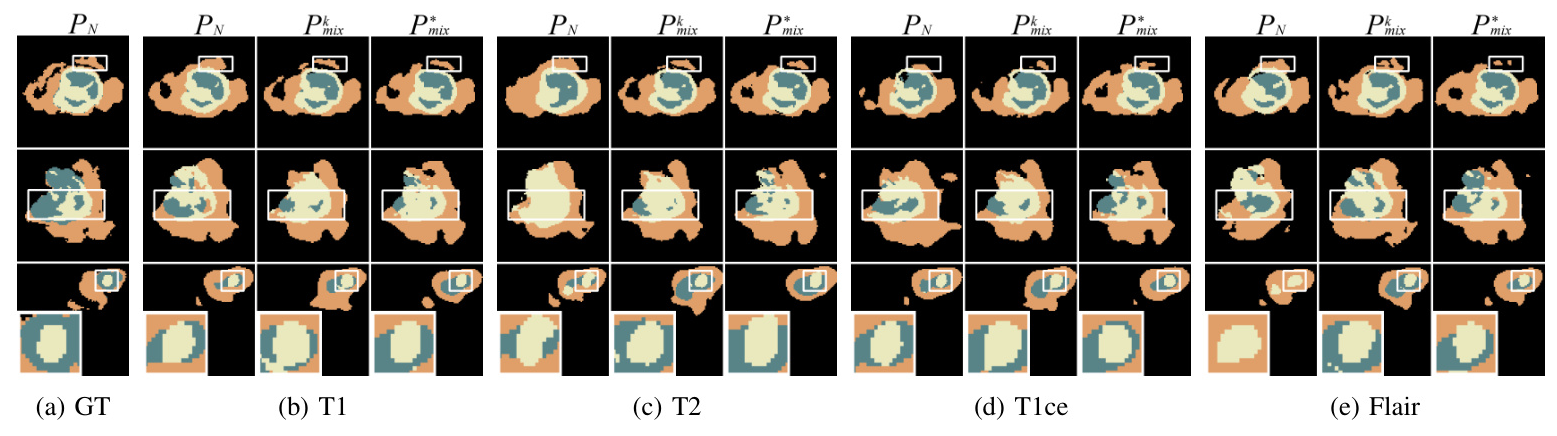
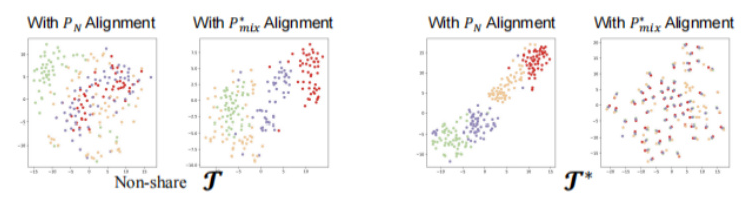
Comparison of Alignment Paradigm Components
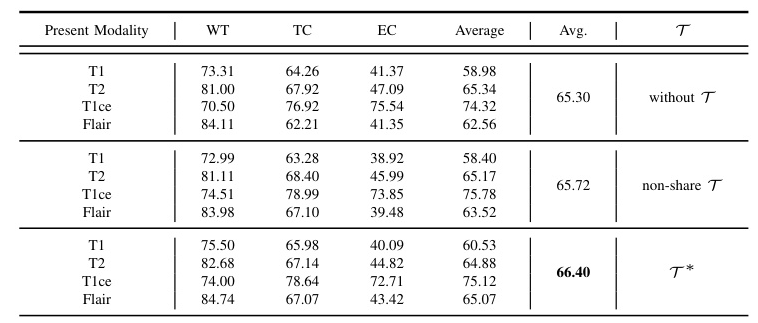
Ablation studies show that the enhanced encoder (T) and the adaptive alignment anchor (P_mix) yield the best performance. The t-SNE visualizations further confirm that MedMAP effectively narrows the gaps between modalities.
Ablation Studies of Hyperparameters
The initialization of P*_mix and the alignment weight parameter are crucial for the convergence and performance of the model. Empirical studies suggest that setting the alignment loss to one-eighth of the original loss yields optimal results.
Overall Conclusion
This paper presents MedMAP, a novel alignment paradigm for brain tumor segmentation with missing modalities. By aligning latent features to a pre-defined distribution, MedMAP effectively minimizes modality gaps and improves segmentation performance. Extensive experiments demonstrate the superiority of MedMAP over several state-of-the-art approaches, making it a promising solution for medical image segmentation tasks with incomplete modalities.