Authors:
Stefano Puliti、Emily R. Lines、Jana Müllerová、Julian Frey、Zoe Schindler、Adrian Straker、Matthew J. Allen、Lukas Winiwarter、Nataliia Rehush、Hristina Hristova、Brent Murray、Kim Calders、Louise Terryn、Nicholas Coops、Bernhard Höfle、Samuli Junttila、Martin Krůček、Grzegorz Krok、Kamil Král、Shaun R. Levick、Linda Luck、Azim Missarov、Martin Mokroš、Harry J. F. Owen、Krzysztof Stereńczak、Timo P. Pitkänen、Nicola Puletti、Ninni Saarinen、Chris Hopkinson、Chiara Torresan、Enrico Tomelleri、Hannah Weiser、Rasmus Astrup
Paper:
https://arxiv.org/abs/2408.06507
Benchmarking Tree Species Classification from Proximally-Sensed Laser Scanning Data: Introducing the FOR-species20K Dataset
Introduction
In recent years, there has been a significant push towards automating the retrieval of key forest variables from various remotely sensed data, with laser scanning technology providing the most detailed and accurate 3D information. Laser scanning and other 3D technologies have demonstrated exceptional capabilities in capturing forest structure, and a variety of scanning platforms exist, suited to a range of target applications. These applications range from large-scale airborne forest management inventories employing airborne laser scanning (ALS) to small-scale surveys aimed at capturing in-situ data and relying on proximally-sensed laser scanning data from various platforms, which we here define as uncrewed aerial vehicle (UAV) laser scanning (ULS), terrestrial laser scanning (TLS), and mobile laser scanning data (MLS).
While the value of laser scanning for increased understanding of the structure and function of forests has been widely acknowledged, its ability to routinely replace traditional ground surveys is limited by several factors. These include the cost and accessibility of sensors, and the significant manual work required for post-processing to retrieve interpretable information. Crucially, standard approaches for large-scale, reliable retrieval of ecologically important forest properties have not yet been agreed upon by the community. The classification of tree species from laser scanning data is a crucial task for effective forest monitoring, management, and assessment of forest functions and ecosystem services.
Deep learning methods have shown promise for the problem of individual tree species classification, but development has largely relied on single datasets from a small number of species in a single ecosystem type. Reliable and robust universal models have not emerged, limiting the use of laser scanning sensors to monitor forests and their ecosystem services without auxiliary ground data.
A review of a substantial body of literature identified key trends and gaps in tree species classification using individual tree point clouds:
- Platform Variation: The variation in platforms used to capture laser scanning data illustrates the broad spectrum of downstream applications and underlines the importance of point cloud-based tree species classification.
- Advancements in State-of-the-Art: Over the past decade, there has been a general improvement in performance, particularly due to the advent of deep learning (DL) methods.
- Geographic, Species, and Ecosystem Diversity in Studies: Most studies have been conducted in mature and relatively simple forest types, predominantly in China and Finland.
- Dataset Size and Diversity: Most studies used small datasets, averaging around 1,500 trees, with limited species diversity.
- Platform and Sensor Agnostic Models: Previous research has been constrained to single laser scanning data modalities, leading to a significant gap in understanding the cross-platform performance of different methods.
- Accessibility of Data and Code: Out of the nineteen studies reviewed, only a few made their datasets publicly available.
Materials
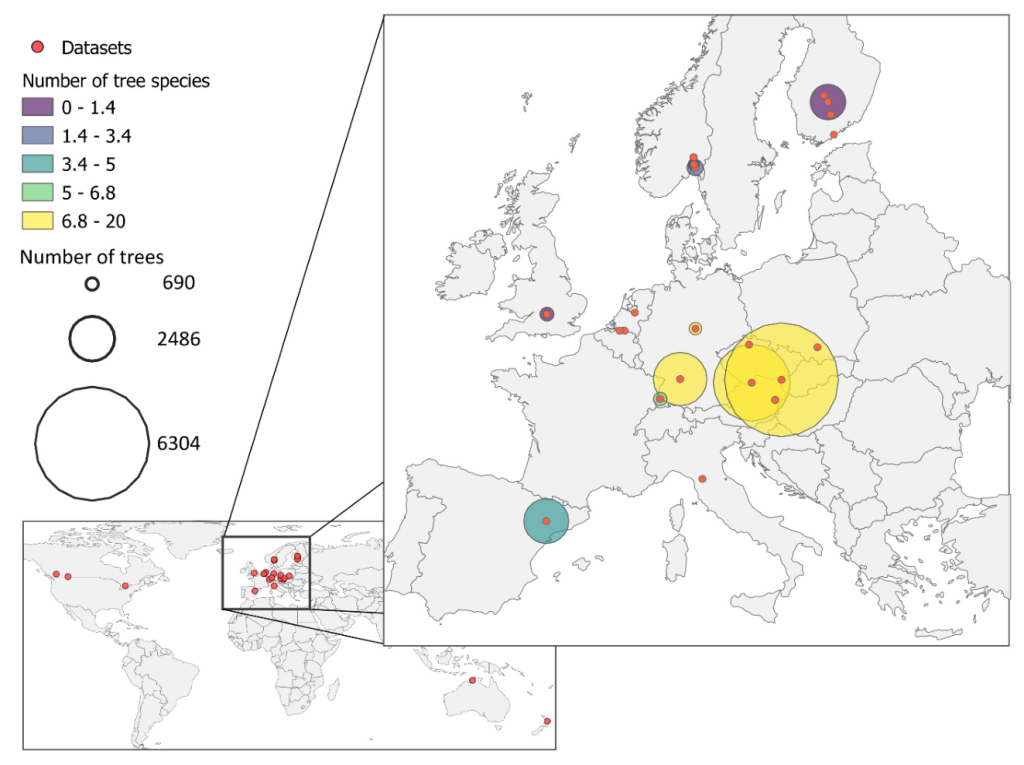
The FOR-species20K dataset was compiled by combining 25 different datasets, composed of either open datasets or in-kind contributions from researchers, primarily in Europe but also in North America and Australia. The basic data unit is individual tree point clouds, ensuring high-quality individual tree segmentation. The final number of trees included in the FOR-species20K dataset was 20,158.
Data Origin
The large majority (90% of the trees) of data in the FOR-species20K were collected within Europe, with additional scattered data collections from Canada, Australia, and New Zealand.
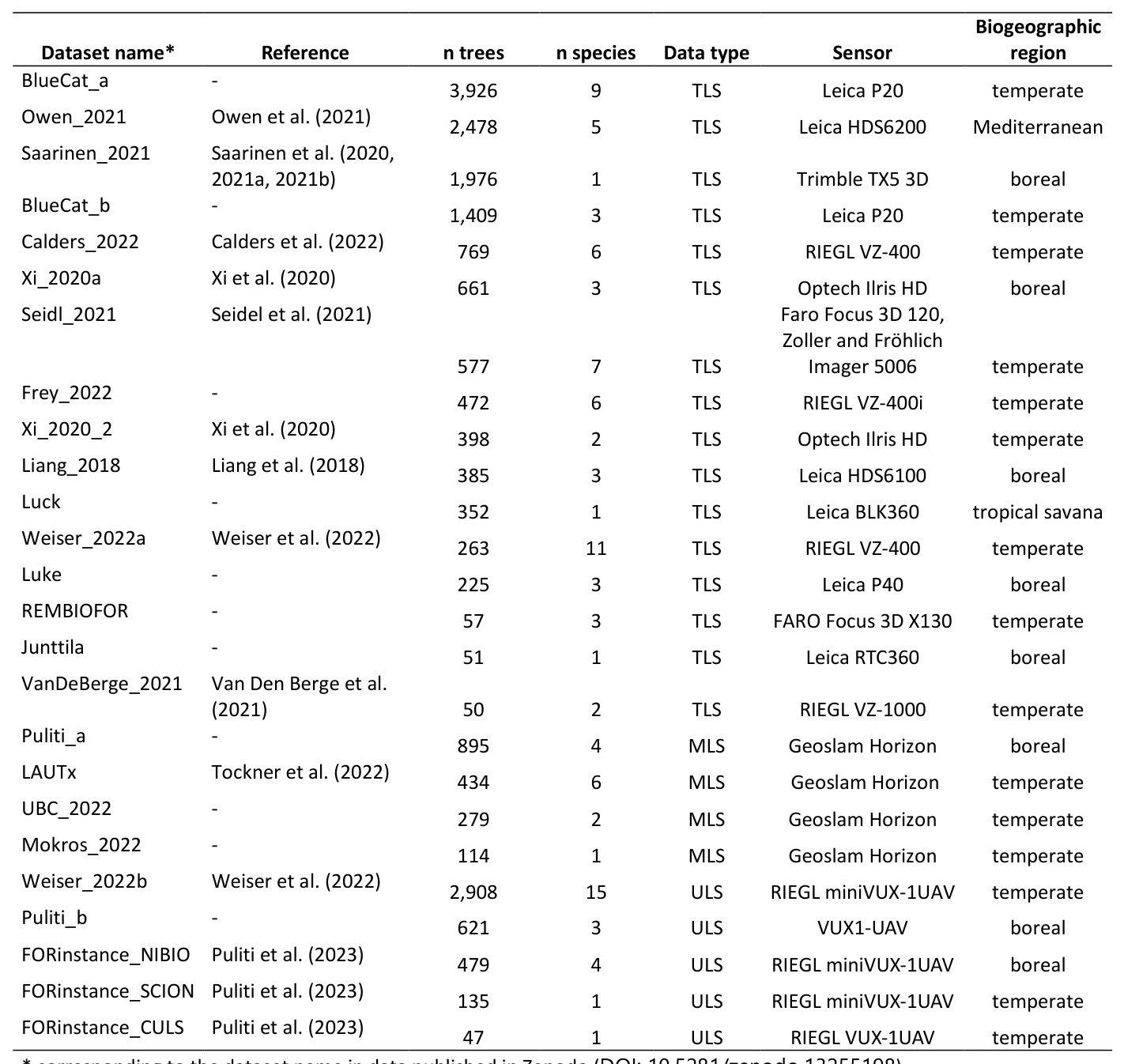
Platforms and Sensors
The FOR-species20K dataset is composed mostly of TLS data (70% of the trees), followed by ULS (22%), and MLS data (8%). The TLS data is characterized by a broader variability in terms of sensors and scanning protocols. MLS and ULS data came from only one type of sensor each.
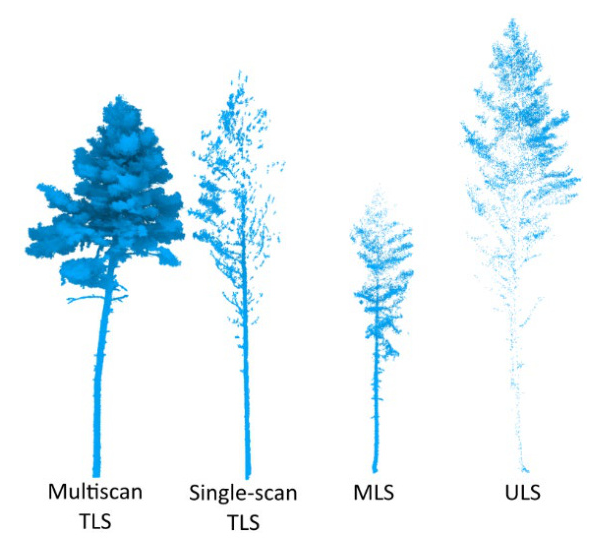
Forest Types and Tree Species
The FOR-species20K data covers the three main forest ecoregions in Europe, with most of the trees from temperate forests (61%), followed by boreal forests (25%) and Mediterranean forests (7%). The dataset includes 33 species of 19 genera, with a realistic abundance distribution reflecting European forest ecosystems.
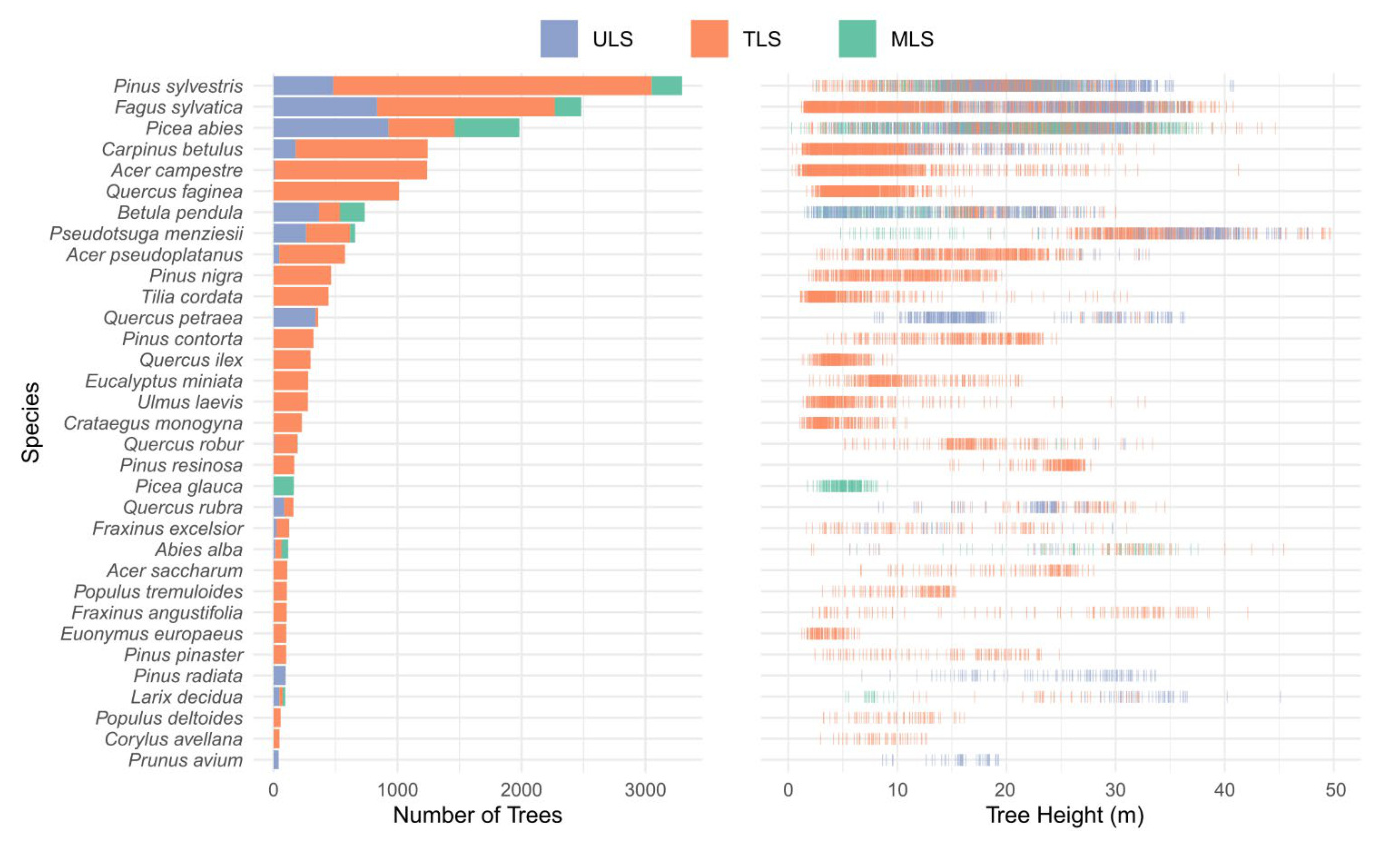
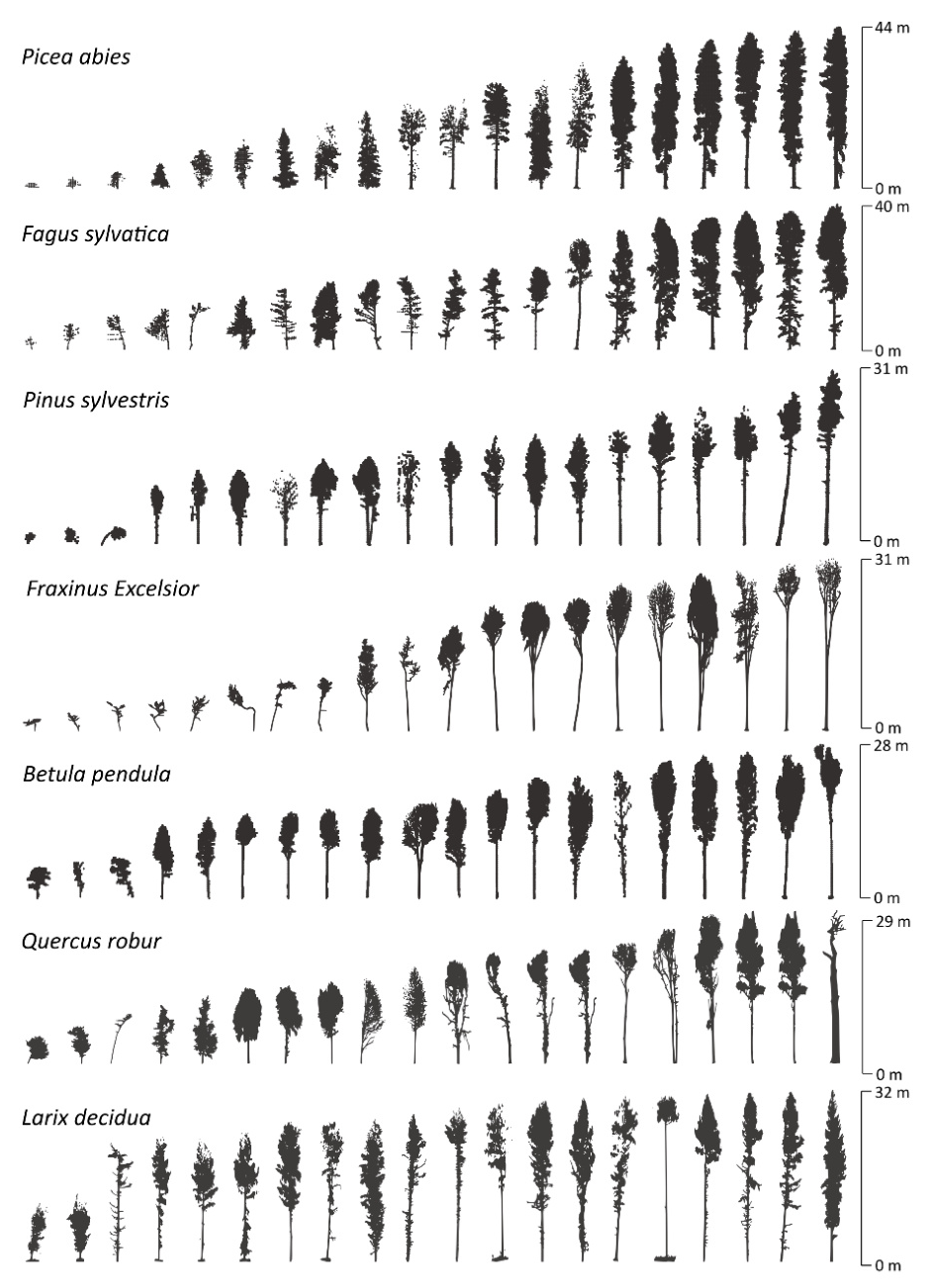
Dataset Variability in Tree Size and Crown Architecture
The FOR-species20K dataset captures significant variation in tree height and crown architecture, encompassing a broad range of tree developmental stages under various growth conditions.
Data Split
To benchmark different classifiers, the full dataset was split into development (90% of the trees) and test set (10% of the trees). The test dataset was selected based on a stratified random sampling aimed at creating an artificially balanced dataset.
Methods
Data Science Competition
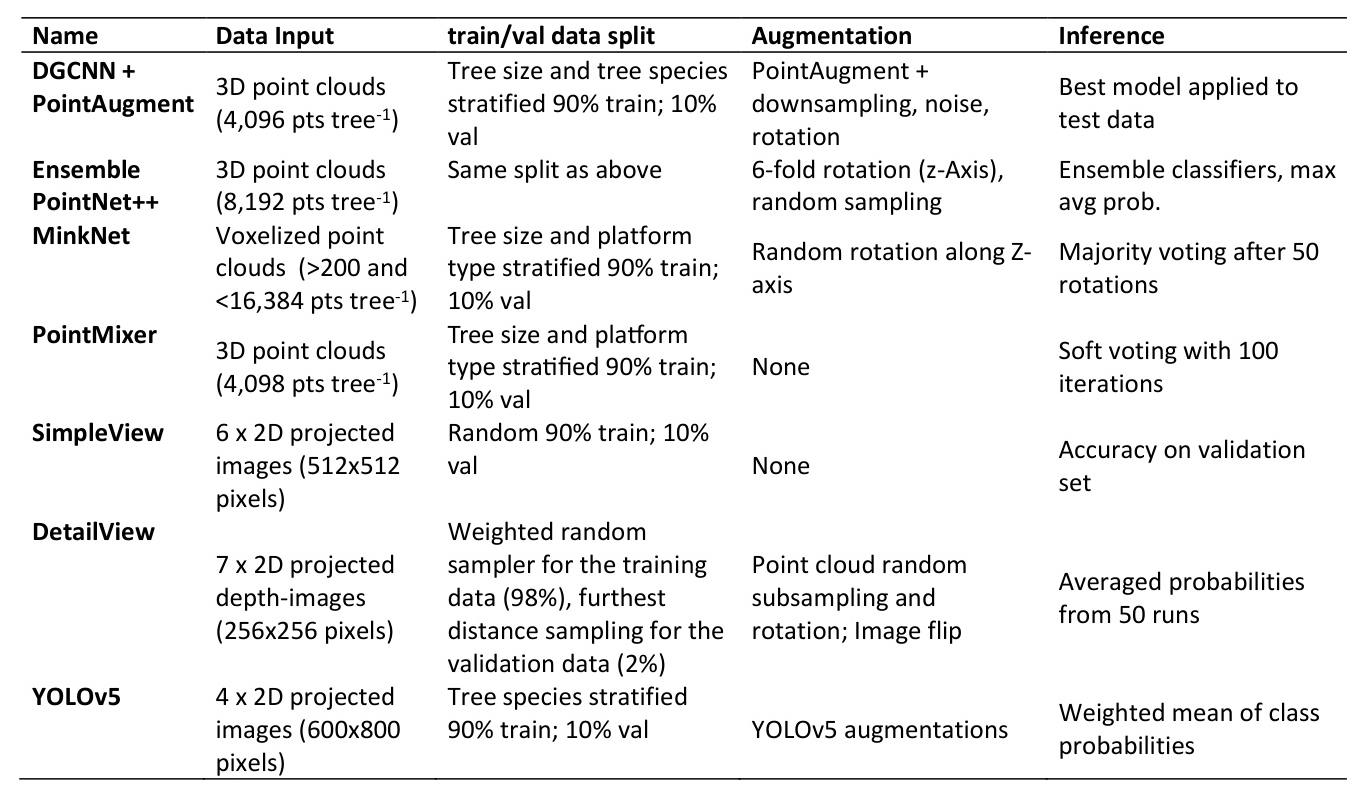
A data science competition was launched to benchmark some of the most common classifiers available in the literature, including some of the latest model architectures. Competitors tested seven different DL methods, including methods operating either directly on 3D point clouds or on 2D images obtained from projecting point clouds.
Point-Cloud Based Methods
- PointAugment and DGCNN: Combines PointAugment, a generative adversarial network (GAN), and the Dynamic Graph Convolutional Neural Network (DGCNN).
- Ensemble PointNet++: Utilizes PointNet++ to extract features directly from 3D point clouds.
- MinkNet: Employs 3D sparse convolutions on voxelized point clouds using the Minkowski Engine framework.
- PointMixer: Blends features within and between point sets, making it effective for tree species classification.
Image-Based Methods
All tested image-based methods relied on multi-view approaches, projecting point clouds onto 2D planes from different viewpoints.
- SimpleView: Uses a multi-view approach with six orthogonal camera projections of point clouds.
- DetailView: Builds upon SimpleView and incorporates dataset balancing by species, tree size, and platform.
- YOLOv5: Applies a modified YOLOv5 architecture and uses orthographic projections colored by point count.
Benchmarking and Metrics
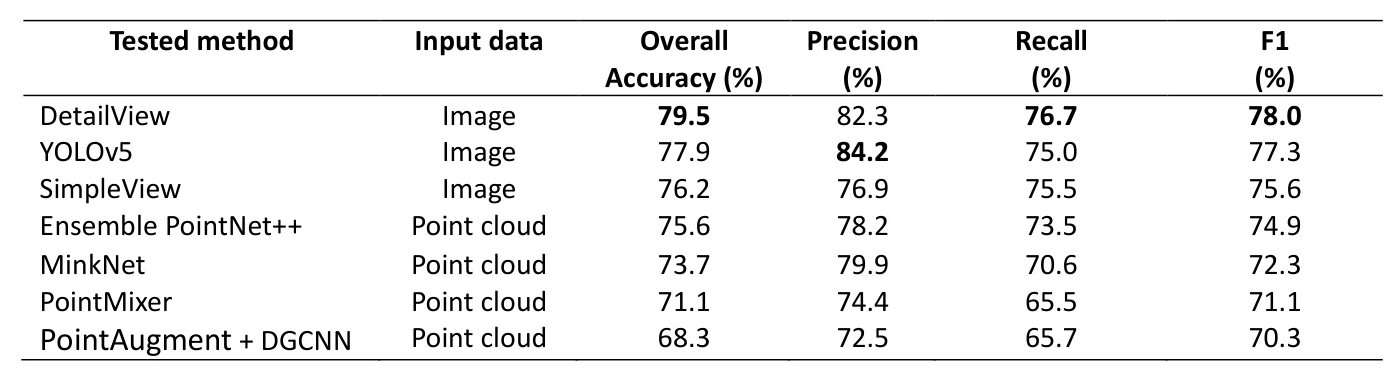
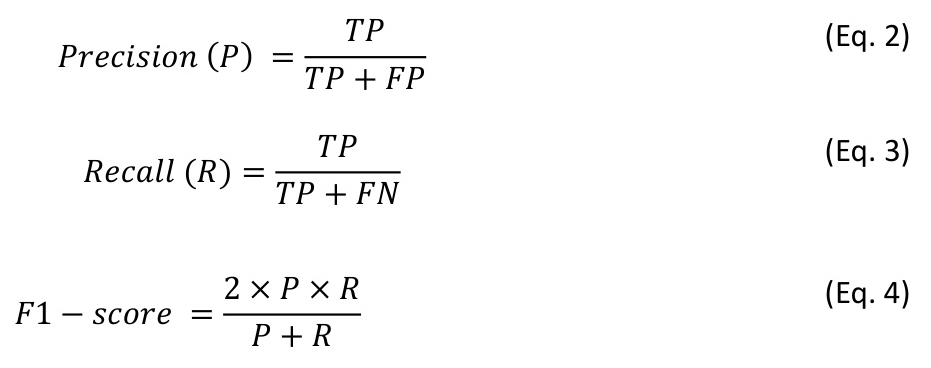
The models were benchmarked in relation to their ability to classify tree species in the unseen test data. Metrics used include overall accuracy (OA), precision, recall, and F1-score.
Results and Discussion
Benchmarking
The leaderboard for the data science competition showed that the best performing model by overall accuracy (OA), recall, and F1-score was DetailView, while YOLOv5 had the best precision. Multi-view image-based methods had higher average OA than point cloud methods.
Accuracy by Acquisition Platform
The assessment of platform-agnostic capabilities revealed that all models performed well regardless of the laser scanning data source. DetailView performed best for MLS and ULS data, but was outperformed by YOLOv5 for TLS data.
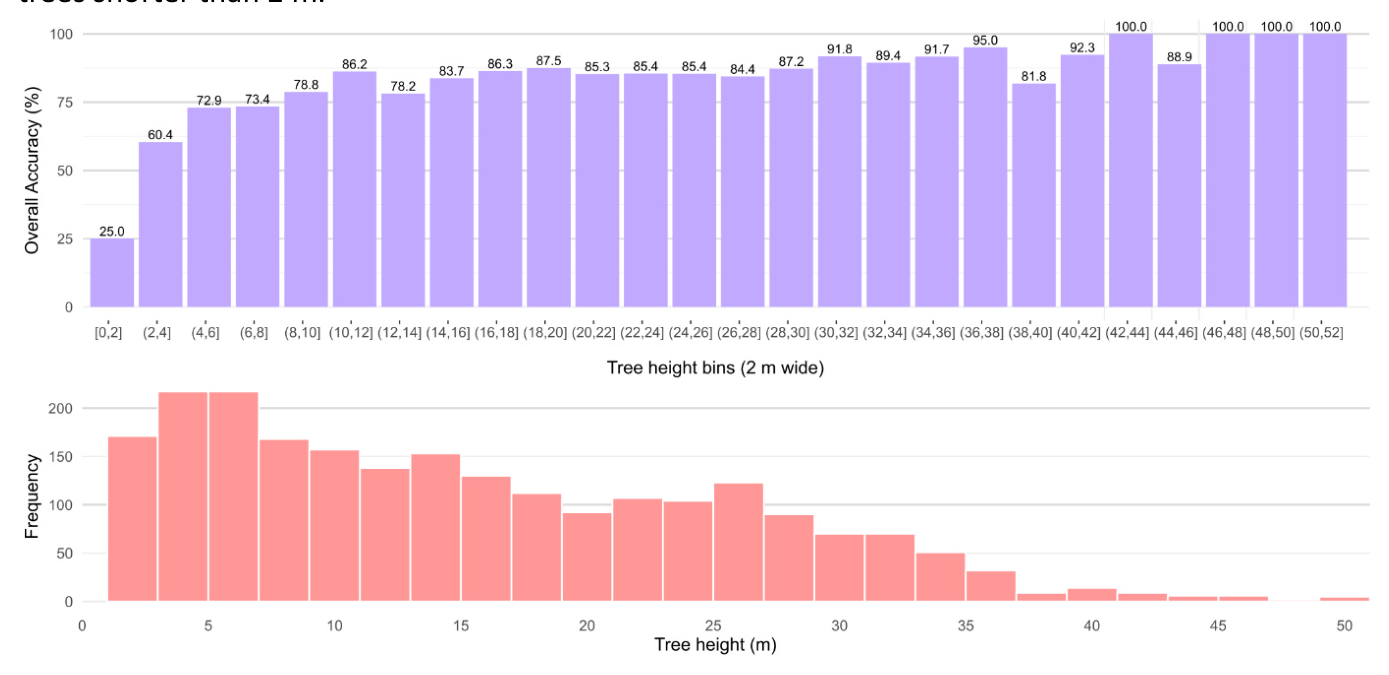
Accuracy by Tree Size
DetailView’s overall accuracy was stable for trees taller than around 8 meters but substantially lower for small trees (< 5 meters in height).
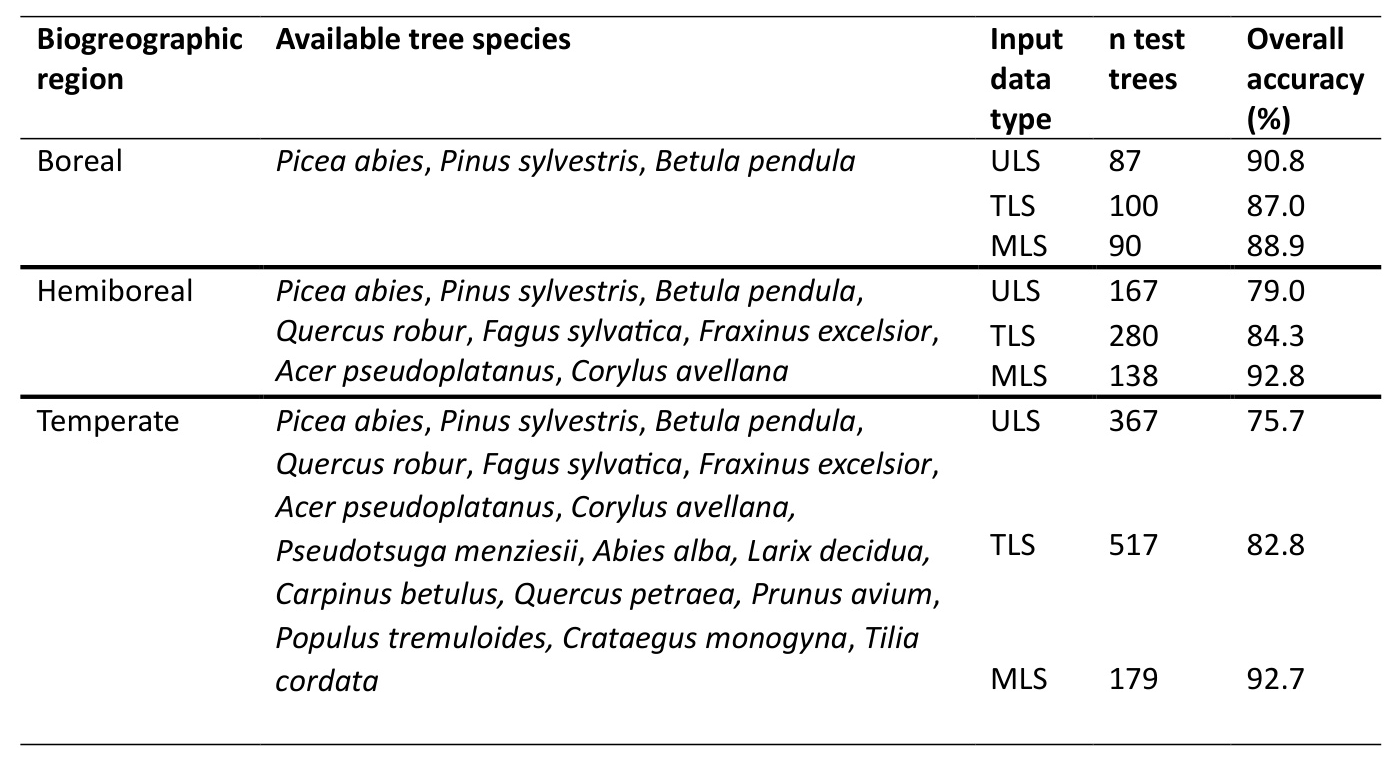
Domain/Application Evaluation
DetailView performed reliably across different biomes, consistently achieving high overall accuracy in species-poor biomes and decreasing as tree species richness increased.
Limitations
Users must be aware of potential limitations, including the incomplete species list, differences in tree segmentation quality, and species representation on different platforms or sensors.
Conclusions
This study addresses the need for benchmarked individual tree species classifiers, contributing to the accurate characterization of forest ecosystems using automated methods relying on proximal laser scanning data. The FOR-species20K dataset represents an important starting point, and further efforts are needed to expand the database with more tree species, increase sample sizes for poorly represented species, and include more trees from underrepresented scanning approaches.
Acknowledgements
This work was supported by various funding agencies and institutions, including the COST Action 3DForEcoTech, the Center for Research-based Innovation SmartForest, UKRI Future Leaders Fellowship, and others.
Data Availability Statement
The FOR-species20K data are openly available at Zenodo, and the code for the different methods benchmarked in this study can be found at the following GitHub repositories: