Authors:
Jingyun Chen、Martin King、Yading Yuan
Paper:
https://arxiv.org/abs/2408.10275
FedKBP: Federated Dose Prediction Framework for Knowledge-Based Planning in Radiation Therapy
Introduction
Radiation therapy is a cornerstone in the treatment of cancer, and the demand for efficient and effective treatment planning is ever-growing. Knowledge-based planning (KBP) has emerged as a promising approach to streamline the planning process and reduce treatment lead time. Central to KBP is the dose prediction, which automatically estimates patient-specific dose distribution for treatment plan evaluation and optimization. However, the challenge of limited training data availability has been a significant hurdle in the development of robust dose prediction models.
Recent advances in deep learning-based dose prediction methods have shown promise, but the need for large and diverse datasets remains a critical issue. Federated learning (FL) has emerged as a solution, enabling medical centers to jointly train deep-learning models without compromising patient data privacy. This study introduces FedKBP, a federated dose prediction framework designed to evaluate the effectiveness of federated learning in KBP.
Related Work
Knowledge-Based Planning (KBP)
KBP has been introduced to improve the efficiency and accuracy of radiation therapy planning. It leverages historical treatment data to predict optimal dose distributions for new patients, thereby reducing the time and effort required for manual planning.
Deep Learning-Based Dose Prediction
Recent studies have explored the use of deep learning models for dose prediction in radiation therapy. These models have demonstrated strong performance but are often limited by the availability of large and diverse training datasets.
Federated Learning (FL)
FL is a distributed machine learning approach that allows multiple institutions to collaboratively train models without sharing raw data. This approach preserves data privacy and security while enabling the development of robust models using diverse datasets.
Research Methodology
Data
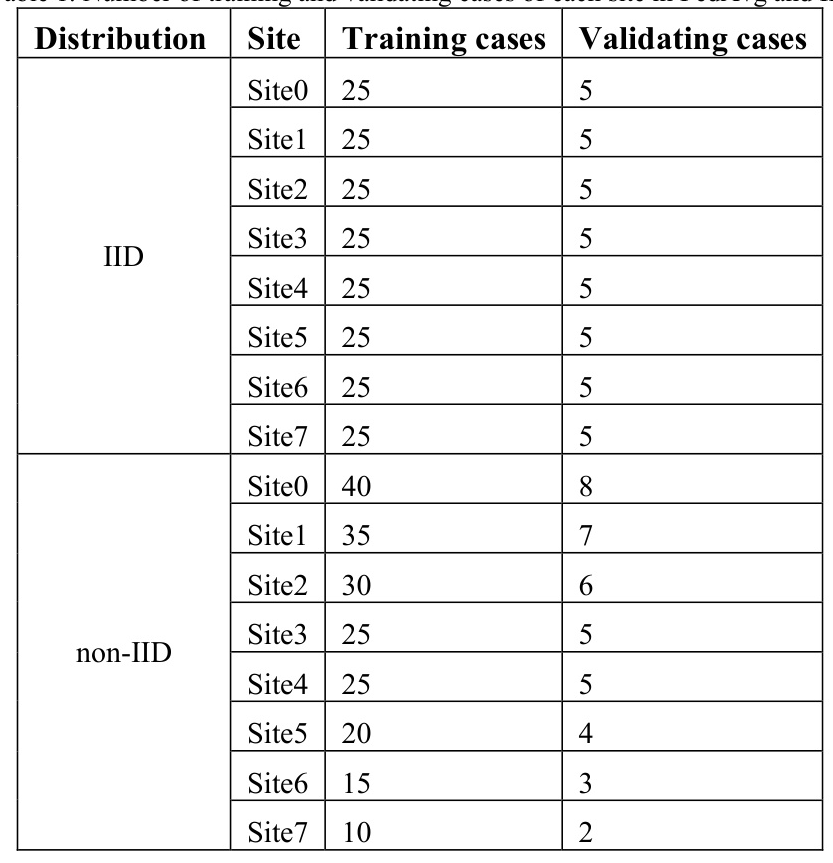
The study employed the public dataset from the OpenKBP grand challenge, which includes 340 cases for head and neck cancer treatment. The dataset was divided into 200 training cases, 40 validating cases, and 100 testing cases. To simulate a distributed training environment, the training and validating cases were randomly divided into 8 groups, representing 8 training sites. Two types of data distributions were implemented: Independent and Identically Distributed (IID) and non-IID.
Dose Prediction Model
The Scale Attention Network (SANet) was implemented for 3D dose prediction. SANet features a dynamic scale attention mechanism that integrates low-level details with high-level semantics from feature maps at different scales. The voxel-wise Mean Absolute Error (MAE) between dose prediction and ground truth was used as the loss function.
Baselines and Evaluation
Three different training methods were evaluated:
1. Pooled Model (PM): Trained with all 8 sites’ training data pooled together.
2. Individual Model (IM): Each site trains with its local data individually without exchanging models.
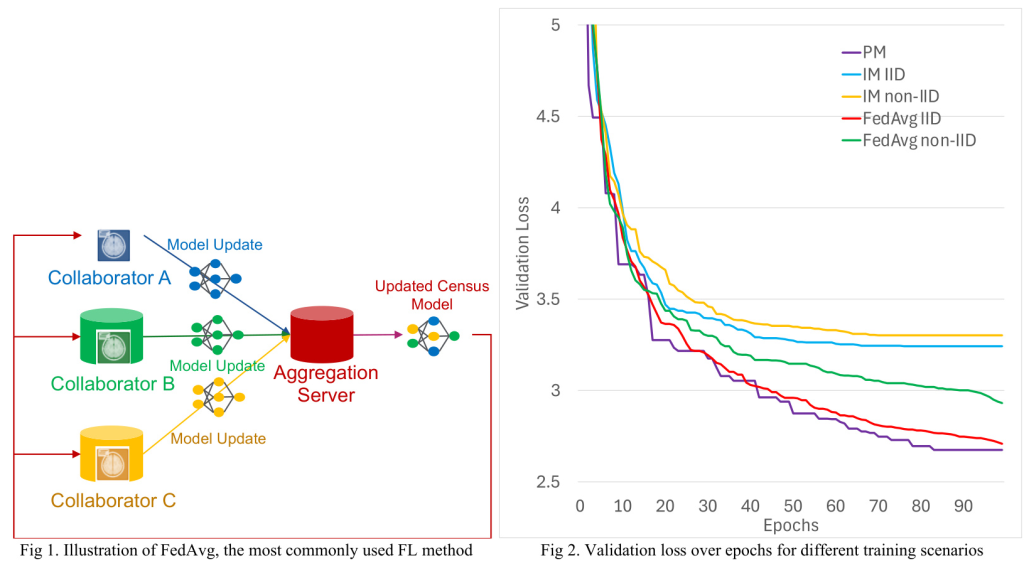
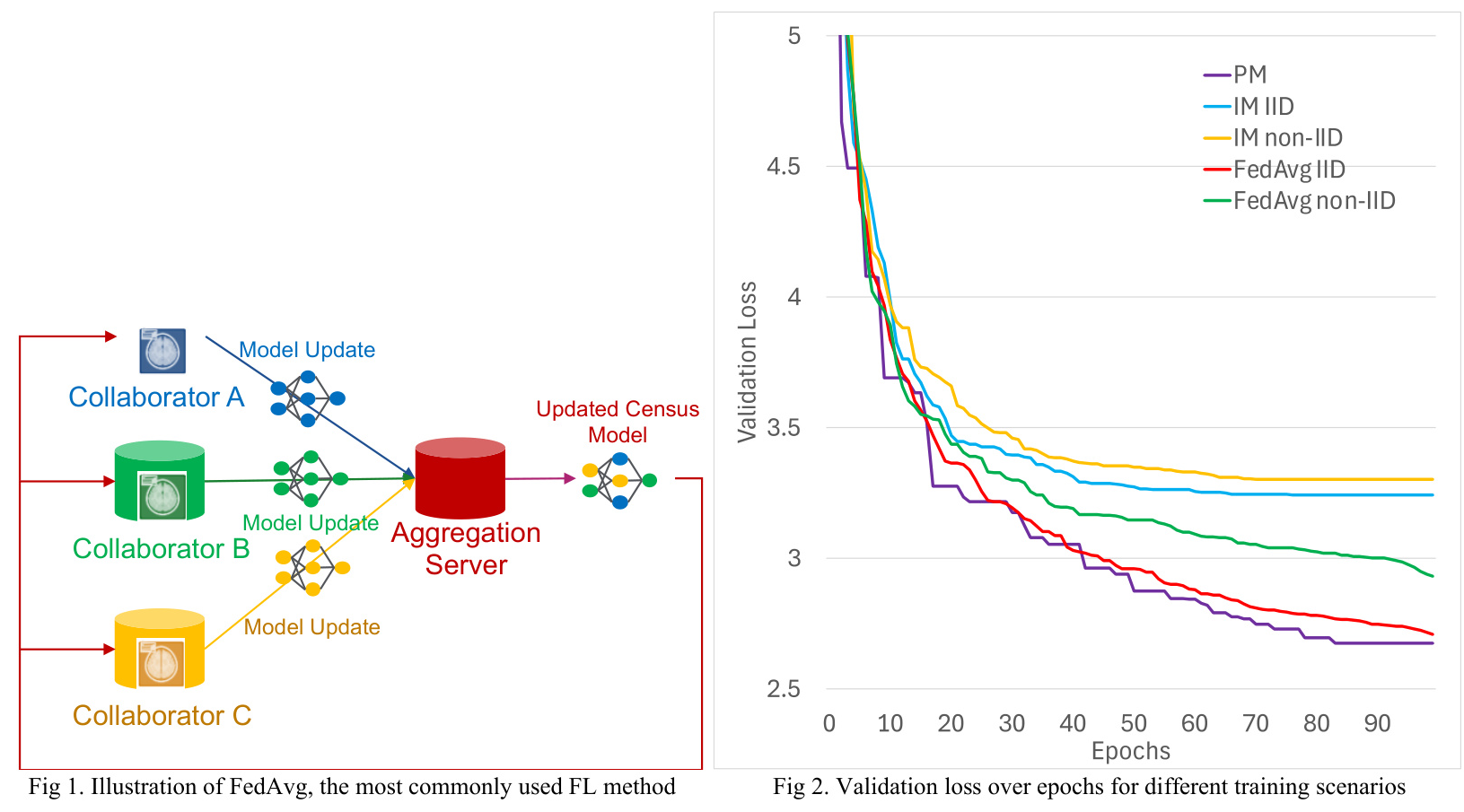
3. FedAvg: Forms a consensus model at each round by weighted average of the local models from all sites.
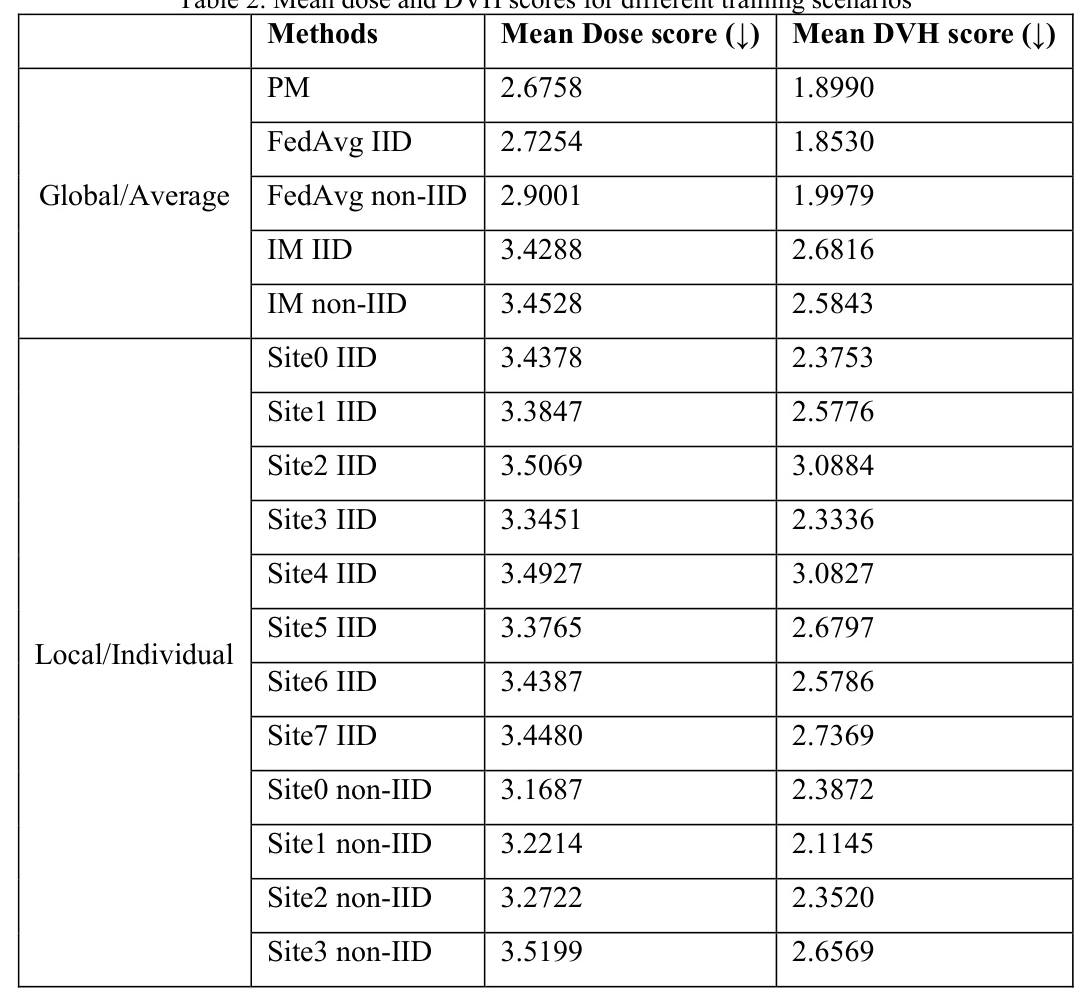
The model performances were assessed using dose score and dose-volume histogram (DVH) score, with smaller values representing higher prediction accuracy and better model performance.
Implementation
All training methods were carried out using the in-house developed FedKBP framework on Nvidia GTX 1080 TI GPUs. Each method was trained for 100 epochs, with each site assigned a separate GPU for training in the case of FedAvg and IM.
Experimental Design
Data Preparation
The OpenKBP dataset was divided into training, validating, and testing sets. The training and validating sets were further divided into 8 groups to simulate distributed training environments. Two types of data distributions (IID and non-IID) were implemented to evaluate the effect of inter-site data variation on model training.
Model Training
The SANet model was trained using three different methods (PM, IM, FedAvg) under both IID and non-IID data distributions. The training process was carried out for 100 epochs, and the model performances were evaluated using dose and DVH scores.
Evaluation Metrics
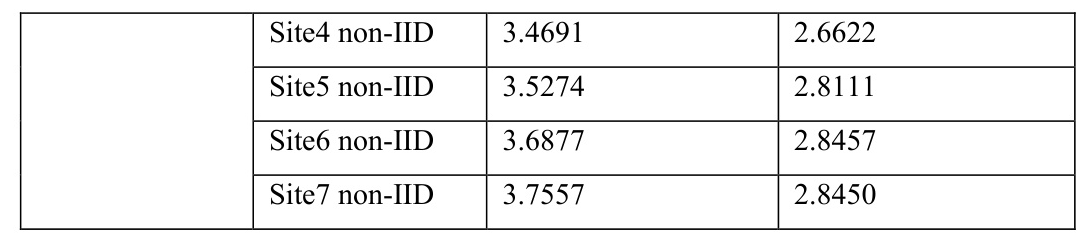
The model performances were assessed using dose score and DVH score, with smaller values indicating higher prediction accuracy. The validation loss over epochs was also monitored to evaluate the model optimization over time.
Results and Analysis
Global/Average Level Performance
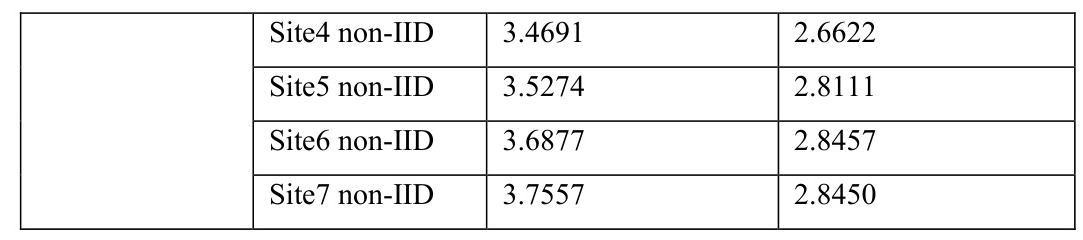
The pooled model (PM) showed high performance on both dose and DVH scores, as expected. FedAvg consistently outperformed IM on both testing scores and data divisions, demonstrating the advantages of federated learning. Notably, the IID FedAvg showed comparable performance to PM, indicating that FL can achieve similar performance to pooled training even with distributed data.
Local/Individual Level Performance
The IID IM showed coherent performances among local sites, while the non-IID IM exhibited variability, with larger sites performing better than smaller sites. This finding highlights the benefit of large datasets and the necessity of collaboration among data contributors for improved performance.
Validation Loss
The validation loss over epochs showed that PM had the fastest decrease and lowest final value among all training scenarios. FedAvg showed faster decrease and lower final values than IM under both IID and non-IID data distributions, reflecting enhanced model optimization in FedAvg.
Overall Conclusion
The study introduced FedKBP, a federated dose prediction framework for knowledge-based planning in radiation therapy. The results underscored federated learning as a promising solution for training dose prediction models over distributed data while preserving data privacy. However, non-IID data distribution poses a challenge to FL, necessitating more sophisticated methods to tackle data variations among participating sites.
Federated learning offers a promising alternative to centralized data-pooling, delivering comparable performance while preserving data privacy. Future work should focus on developing advanced FL methods to address the challenges posed by non-IID data distributions and further enhance model performance.
Acknowledgements
This work is supported by a research grant from Varian Medical Systems, UL1TR001433 from the National Center for Advancing Translational Sciences, and R21EB030209 from the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health, USA. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This research has been partially funded through the generous support of Herbert and Florence Irving/the Irving Trust.