Authors:
Ievgeniia A. Tiukova、Daniel Brunnsåker、Erik Y. Bjurström、Alexander H. Gower、Filip Kronström、Gabriel K. Reder、Ronald S. Reiserer、Konstantin Korovin、Larisa B. Soldatova、John P. Wikswo、Ross D. King
Paper:
https://arxiv.org/abs/2408.10689
Introduction
Background
The field of artificial intelligence (AI) has made significant strides in automating scientific research through the development of “Robot Scientists.” These systems autonomously generate hypotheses, design and conduct experiments, interpret results, and iterate the process. Previous iterations, such as ‘Adam’ and ‘Eve,’ have demonstrated the potential of AI in functional genomics and early-stage drug development, respectively. The latest advancement in this domain is the Genesis project, which aims to automate systems biology research, particularly focusing on eukaryotic cells like yeast (S. cerevisiae).
Problem Statement
Despite the advancements in high-throughput methods, the complexity of systems biology models, which involve thousands of interacting components, remains a significant challenge. Traditional human-based scientific methods struggle to keep up with the intricate and vast nature of these models. Genesis aims to address this by automating the entire research cycle, thereby increasing efficiency and reducing costs.
Related Work
Previous Robot Scientists
- Adam: The first Robot Scientist to autonomously discover novel scientific knowledge, focusing on the functional genomics of yeast.
- Eve: Designed for early-stage drug development, Eve’s AI-driven approach outperformed standard drug screening methods and made significant discoveries, such as identifying triclosan as an inhibitor for malaria-causing parasites.
Challenges in Systems Biology
Systems biology models are incredibly complex, involving thousands of genes, proteins, and small molecules. Traditional high-throughput methods are insufficient as they are not hypothesis-led. The complexity of these models makes them beyond human intuitive understanding, necessitating the use of AI tools for efficient model refinement and experiment generation.
Research Methodology
Genesis Architecture
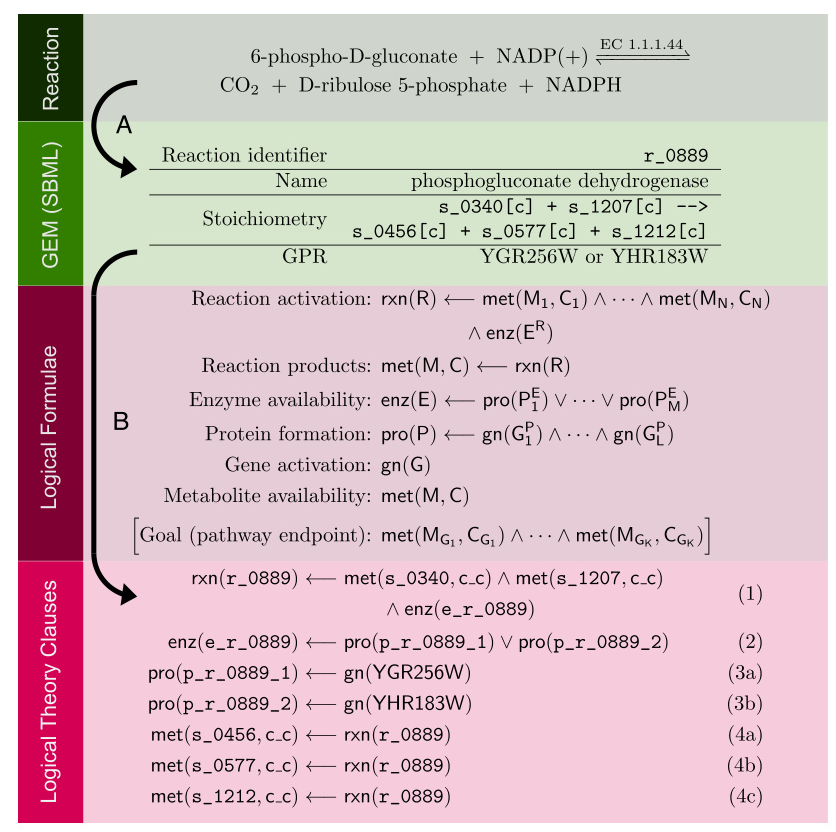
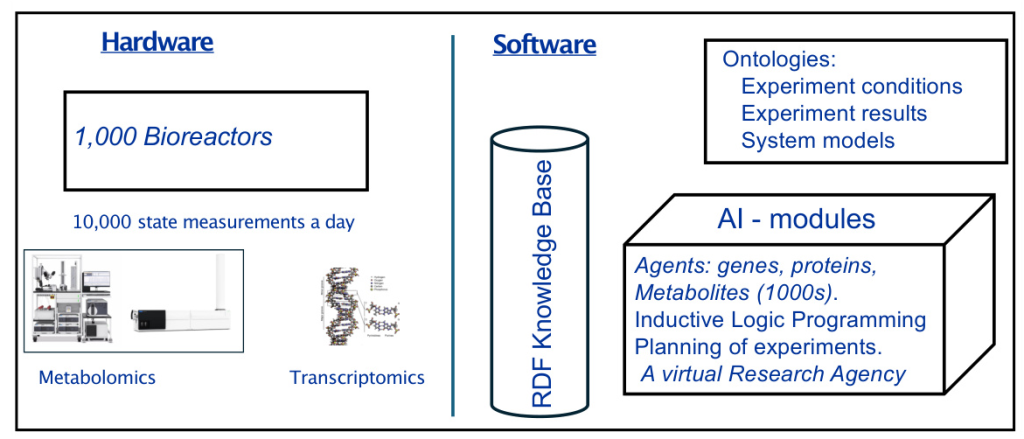
Genesis is designed to automatically improve complex scientific models with thousands of interacting components. The system aims to execute up to one thousand hypothesis-led closed-loop cycles of experiments per day. Each cycle involves hypothesis formation, experiment planning, laboratory execution, and results interpretation.
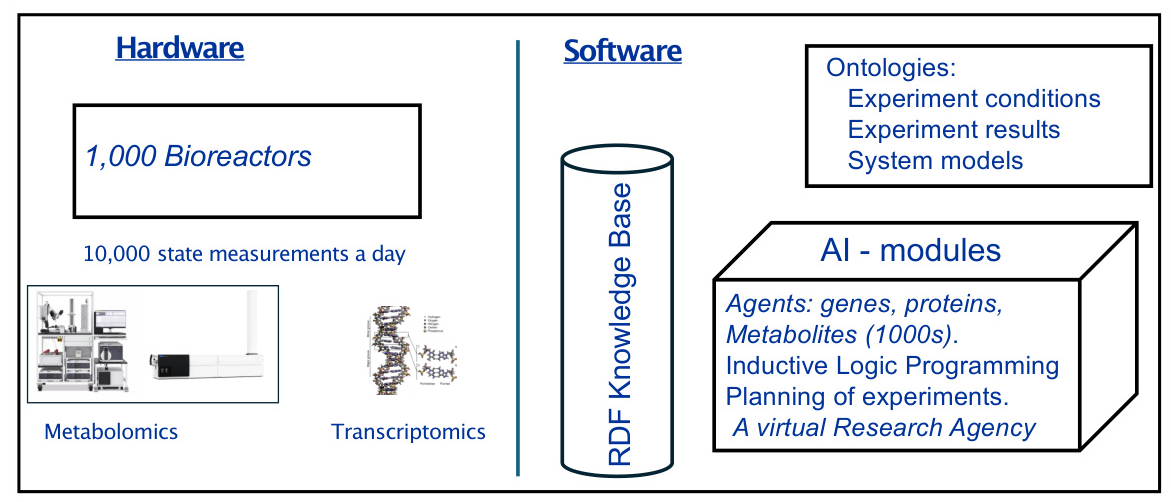
Core Components
- Hardware: The system includes one thousand computer-controlled µ-bioreactors, capable of running in batch, fed batch, or continuous mode.
- Mass Spectrometry: Integration of high-throughput mass spectrometry (IM-MS) for detailed metabolic and transcriptomic analysis.
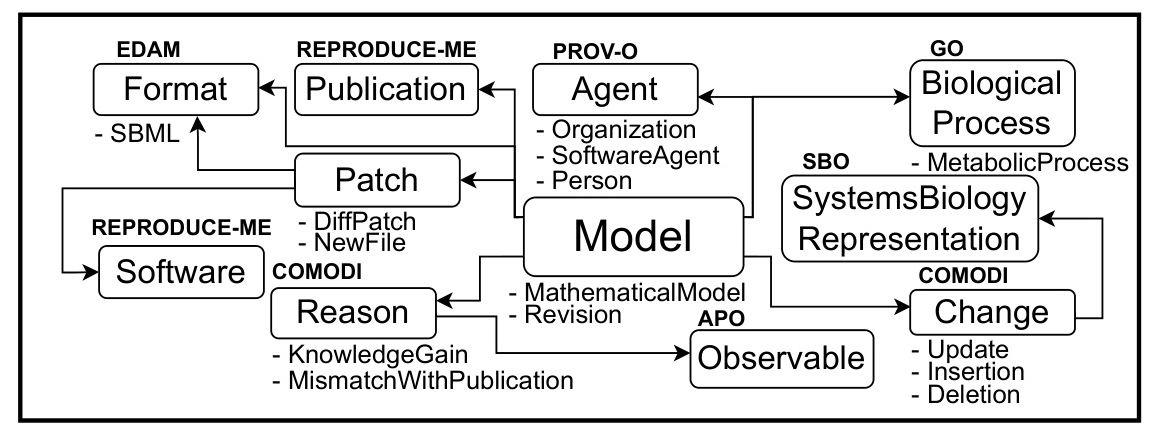
- Ontologies and Databases: Development of Genesis-DB and RIMBO for structured data storage and model revisions.
- Bioinformatics: Utilization of relational learning and inductive logic programming for model improvement.
Experimental Design
Hardware Setup
The hardware setup includes a micro-fluidic system with one thousand µ-bioreactors, arranged in groups of 48. Each bioreactor can be configured in real-time to explore a wide range of biological conditions. The observables from these experiments include growth rate, metabolic analysis, and comprehensive gene expression levels.
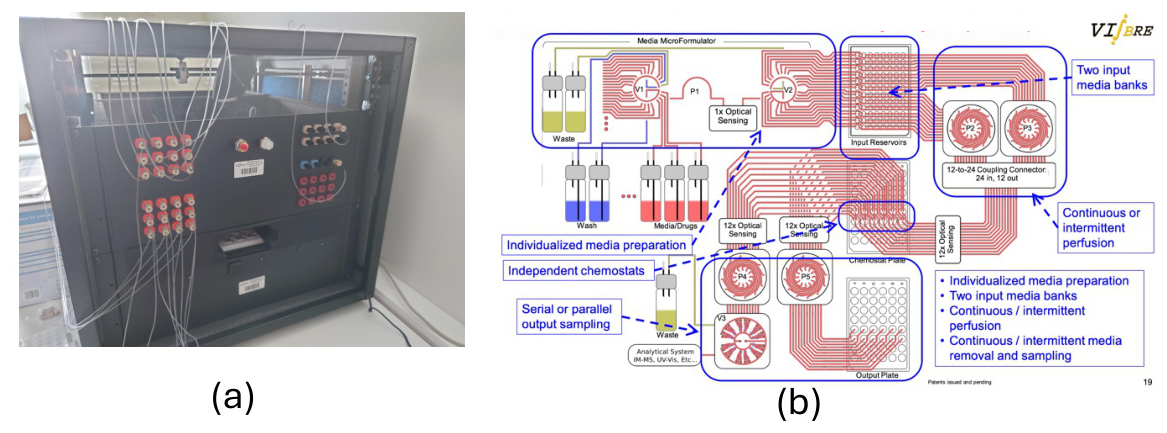
Mass Spectrometry Integration
The mass spectrometry platform, integrated with laboratory automation, enables up to 10,000 measurements a day. The AutonoMS system automates the running, processing, and analysis of high-throughput mass spectrometry experiments.

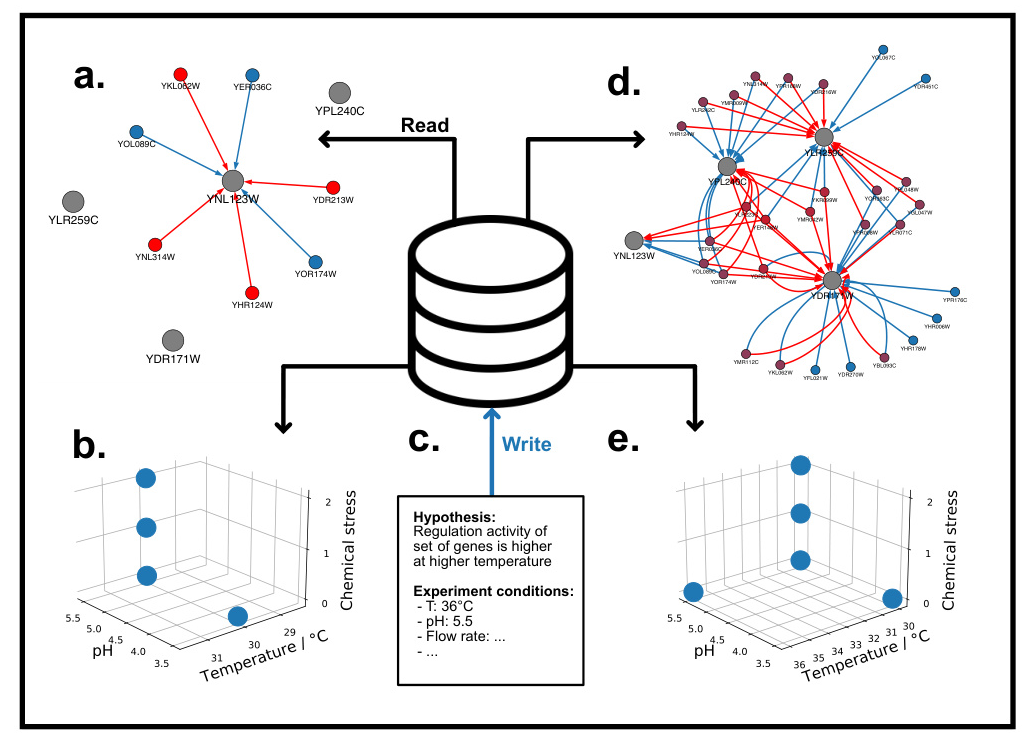
Ontologies and Databases
Genesis-DB supports the research lifecycle by modeling yeast gene regulation and guiding future hypotheses. RIMBO records and describes iterative improvements to models, enabling systematic model revisions.
Bioinformatics Studies
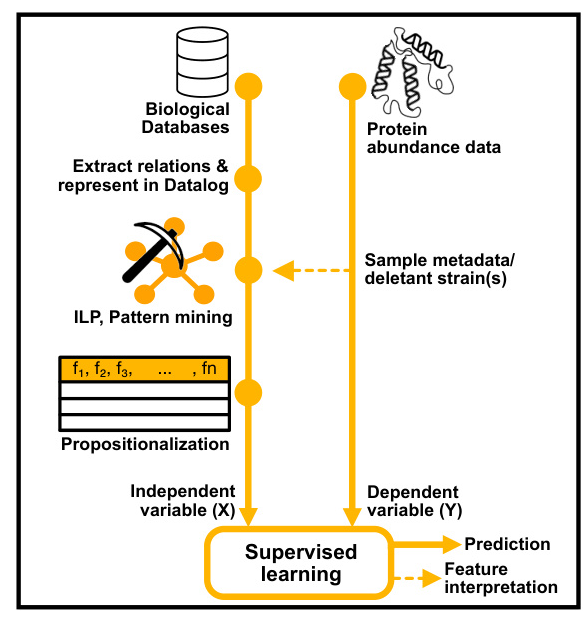
Two bioinformatics studies were conducted to demonstrate the utility of the ontologies and databases. The first study used untargeted metabolomics for functional discovery, while the second focused on proteomics to predict protein abundances.
Results and Analysis
Genesis Hardware
The initial 12 µ-bioreactor system has been successfully implemented, with plans to scale up to 48 units. The flexibility of the µ-bioreactors allows for a wide range of experiments, providing highly informative observables.
Mass Spectrometry
The integration of the Agilent RapidFire and 6560 IM-MS system has demonstrated a high rate of measurements, significantly enhancing the information constraints on modeling.
Ontologies and Databases
Genesis-DB and RIMBO have proven effective in supporting AI-driven discovery and systematic model revisions. The databases enable reasoning about past experiments and planning future ones.
Bioinformatics
The bioinformatics studies validated the use of untargeted metabolomics and proteomics for functional discovery and protein abundance prediction. The relational learning approach provided explainable predictive relationships between protein abundances, function, and phenotype.
Overall Conclusion
The Genesis project represents a significant advancement in the automation of systems biology research. By integrating advanced hardware, mass spectrometry, ontologies, and bioinformatics, Genesis aims to revolutionize the efficiency and cost-effectiveness of scientific research. The project’s success will pave the way for more complex and comprehensive models, ultimately contributing to advancements in medicine, agriculture, and biotechnology.