Authors:
Yusen Wu、Hao Chen、Alex Pissinou Makki、Phuong Nguyen、Yelena Yesha
Paper:
https://arxiv.org/abs/2408.08456
Introduction
Distributional drift, also known as dataset drift, in medical imaging refers to changes in data distribution over time, which can significantly affect the performance of machine learning models used for diagnostic purposes. This drift may result from various factors, including alterations in imaging equipment, differences in imaging protocols, variations in patient demographics, or updates in image preprocessing techniques. Detecting and managing drift is critical in the medical field to ensure that models remain accurate and reliable. Ignoring drift can lead to incorrect diagnoses or suboptimal treatment recommendations, thereby potentially compromising patient care. Therefore, continuous monitoring and adaptation of models are essential to maintain their effectiveness in dynamic clinical environments.
Despite significant advancements in data drift detection within medical imaging, current methodologies exhibit several limitations that must be addressed to improve their effectiveness. We discuss the limitations of L1, L2, and L3 methods as follows:
- L1: Analyzing the distribution of image data is particularly challenging due to its high-dimensional characteristics. Unlike numerical data, where statistical properties like mean and variance can be straightforwardly computed and compared, image data necessitates specialized feature extraction and dimensional reduction techniques to perform meaningful distribution analysis.
- L2: Abnormal datasets can introduce bias in drift detection, which can often be mitigated without compromising the overall data quality. However, efficiently identifying and isolating these abnormal datasets is essential to preserve data integrity. Such abnormalities may arise from data entry errors, sensor malfunctions, or even malicious activities. Implementing effective and efficient anomaly detection techniques is critical for pre-processing data before applying drift detection methods.
- L3: Another limitation is the absence of real-time processing solutions for handling abnormal data, especially during the preprocessing stage. This process often relies on hospital experts to manually label and remove invalid data before training, which significantly increases the cost and complexity of model development. When datasets contain substantial noise or inconsistencies, the drift detection process may incorrectly identify or overlook drifts, thereby compromising the reliability of the analysis.
To overcome these limitations, we fine-tuned a pre-trained model specifically optimized for feature extraction from the target images. We utilized the breast cancer dataset from MetaStar and applied fine-tuning techniques to the Vision Transformer (ViT) model as an example for pre-train model selection. The selection of ViT was based on its superior accuracy post-fine-tuning, as demonstrated by our experimental results in the evaluation section.
Background
Distributional Drift (Dataset Shift) in Medical Images
Distributional drift, also known as dataset shift, refers to changes in the statistical properties of data between the training and deployment phases of machine learning models. In medical imaging, this drift can manifest as differences in pixel intensities, textures, or other visual characteristics between the images used for training and those encountered during real-world application. This phenomenon is particularly prevalent in medical imaging due to the continuous evolution of imaging technologies, variations in imaging equipment, and changes in patient demographics. Such variations can result in significant differences between the training data and new data, challenging the model’s ability to make accurate predictions.
Causes of Distributional Drift in Image Data
Distributional drift in image data can occur due to various factors related to changes in data, environmental conditions, or the image acquisition process. One common cause is variations in lighting conditions, which can occur due to different times of day, weather conditions, or light sources, leading to alterations in brightness and contrast that affect the data distribution. Changes in imaging devices, such as switching cameras or scanning devices, inconsistent calibration, lens aging, or fluctuations in sensor performance, can also result in changes to image quality and subsequently impact the data distribution. Another contributing factor is subject variations, where changes in the shape, posture, or size of the subject being imaged can lead to shifts in the feature distribution of the images. Additionally, modifying the resolution or size of images can alter the distribution of image features, potentially causing drift. Different devices or sensors capturing the same scene may record varying levels of detail, leading to distributional drift. Finally, images taken from different angles of the same subject can introduce changes in viewpoint, resulting in shifts in the data distribution.
Proposed Methods
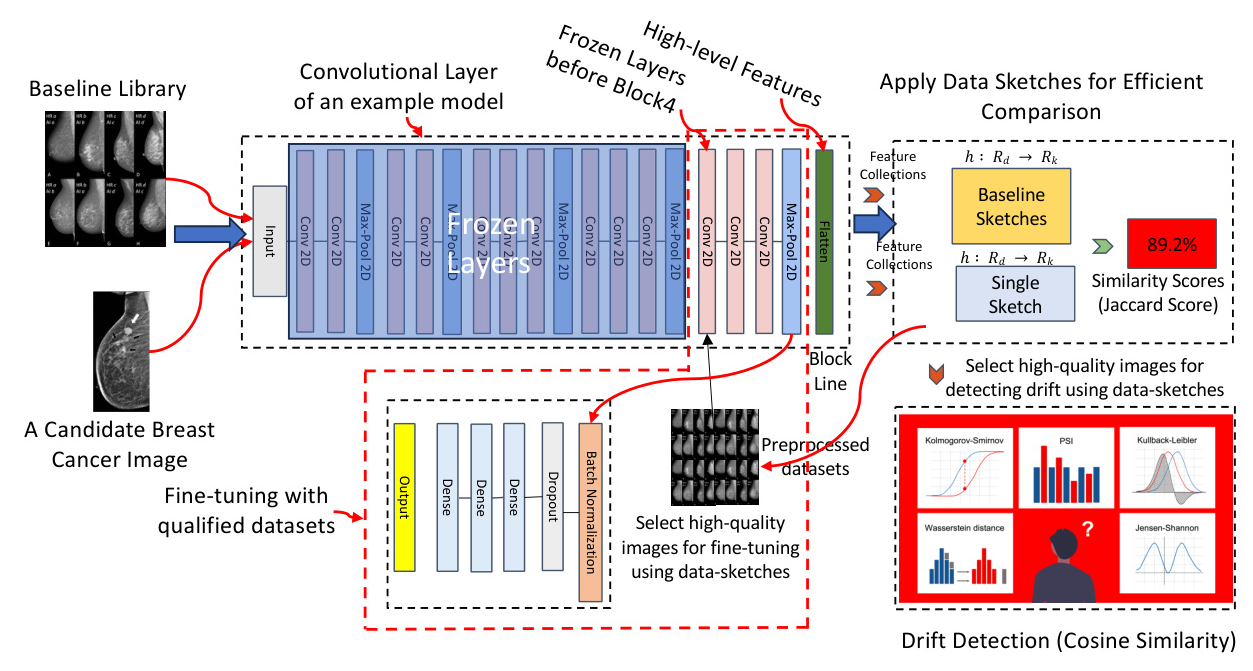
To effectively address the challenges of distributional drift in medical imaging and enhance the accuracy of feature extraction, we propose a comprehensive solution that combines advanced fine-tuning techniques with real-time anomaly detection using data sketching, as shown in Fig. 2. This approach not only optimizes model performance but also ensures the reliable detection of subtle changes in data distribution within dynamic clinical environments.
Fine-tuned Pre-trained Model for Feature Extractions
Our method leverages a pre-trained deep learning model for feature extraction, capitalizing on its ability to encode rich feature representations from input data. Pre-trained models, trained on extensive datasets, offer the advantage of transfer learning, allowing the knowledge gained from large-scale data to be applied to specific tasks with limited data availability. This approach utilizes the pre-trained model within the context of our problem domain to enhance feature extraction.
- Data Preparation: The first step in fine-tuning a model for medical image drift detection involves thorough data preparation. This process includes selecting a qualified dataset, pre-processing the images, and applying data-sketches to improve the data quality.
- Layer Freezing and Fine-Tuning: To preserve the general features learned during pretraining, some of the layers from the pre-trained model are frozen, not updated during the fine-tuning process. Only the final few layers, which are responsible for task-specific features, are fine-tuned.
- Loss Function: The loss function used for training is the Binary Cross-Entropy (BCE) loss, which is suitable for binary classification tasks.
- Optimization Algorithm: We use mini-batch stochastic gradient descent employing the Adam optimizer for training, which adjusts the learning rate for each parameter individually based on estimates of lower-order moments.
- Training Setup: The model is fine-tuned over a series of epochs, and we set it as 20 as the learning curve shows the convergence, which is also depending on the dataset size and complexity.
Data Sketches for Real-time Anomaly Detection
Data sketches are efficient probabilistic data structures that provide approximate representations of large datasets. These structures allow for scalable computations of various statistical properties without requiring the entire dataset to be stored or processed at once. In this subsection, we will delve into the mathematical foundation of data sketches and explain our anomaly detection steps. We summarized our method in Algorithm 1.
- Feature Extraction and Hashing: Given a set of images, we first extract feature vectors from each image using a pre-trained model.
- Constructing the Baseline Library: The baseline library is constructed by aggregating the hashed feature vectors of a representative set of CT-scan images.
- Integration with Real-time Feature Extraction: Our approach seamlessly integrates data sketches with feature extraction to enhance the accuracy and efficiency of drift detection.
- Real-time Anomaly Detection: For an incoming image, we extract its feature vector and hash it to obtain the corresponding data sketch. To detect data drift, we compare the distribution of the new data sketch with the baseline library using the Jaccard similarity.
Similarity Comparisons for Drift Detection
- Kolmogorov-Smirnov (KS) Statistic: The KS statistic is used to compare the empirical cumulative distribution functions (CDFs) of the baseline library and the new data sketch.
- Cosine Similarity: Cosine similarity is a useful metric for comparing two medical images by evaluating the similarity between their feature vectors.
Sensitivity Evaluation via Incremental Noise Introduction
To assess the sensitivity of cosine similarity in detecting data drift, we systematically introduced incremental noise into our experimental dataset and monitored the similarity metric. Specifically, Gaussian noise was added in 10-20% increments to simulate real-world disturbances in medical imaging data, such as fluctuations in image acquisition or processing conditions.
Experimental Results
Experimental Datasets
In our research, we utilize the MedMNIST dataset as a benchmark and 9,987 breast cancer images for fine-tuning. The MedMNIST is an extensive repository of standardized biomedical images specifically created to support the development and assessment of machine learning algorithms in medical imaging. MedMNIST mirrors the structure of the MNIST dataset but is significantly larger and specifically adapted for biomedical purposes. It comprises ten distinct 2D datasets and eight 3D datasets. The ten datasets in MedMNIST encompass a range of medical conditions and imaging techniques. For our analysis, we have selected eight common 2D datasets as benchmarks including PathMNIST, BloodMNIST, OrganMNIST, ChestMNIST, DermaMNIST, BreastMNIST, OCTMNIST, and PneumoniaMNIST.
Evaluation of Model Accuracy
We evaluated the performance of five distinct models—Customized Model, VGG16, ResNet50, ResNet152, and ViT (patch16)—on breast cancer image classification, as shown in Table I. The primary goal was to assess the effectiveness of fine-tuning when applied to pre-trained models in comparison to a baseline CNN model trained from scratch.
| Model | Accuracy | Fine-Tuned Accuracy |
|———————-|———-|———————|
| Customized Model | 78.7% | N/A |
| ResNet50 | N/A | 66.9% |
| ResNet152 | N/A | 66.7% |
| VGG16 | N/A | 92.2% |
| Vision Transformer(ViT) | N/A | 99.11% |
Evaluation of Drift Detection
Baseline 1 (without data-sketches and fine-tuning)
From the experimental results shown in Fig. 3, it is evident that significant data drift starts to emerge around the sixth month. This drift is reflected by the noticeable increase in the Kolmogorov-Smirnov (KS) statistic across most of the MNIST dataset variants, such as PathMNIST, BloodMNIST, and others. This suggests that changes in the underlying data distribution begin at this point, which could negatively impact model performance.
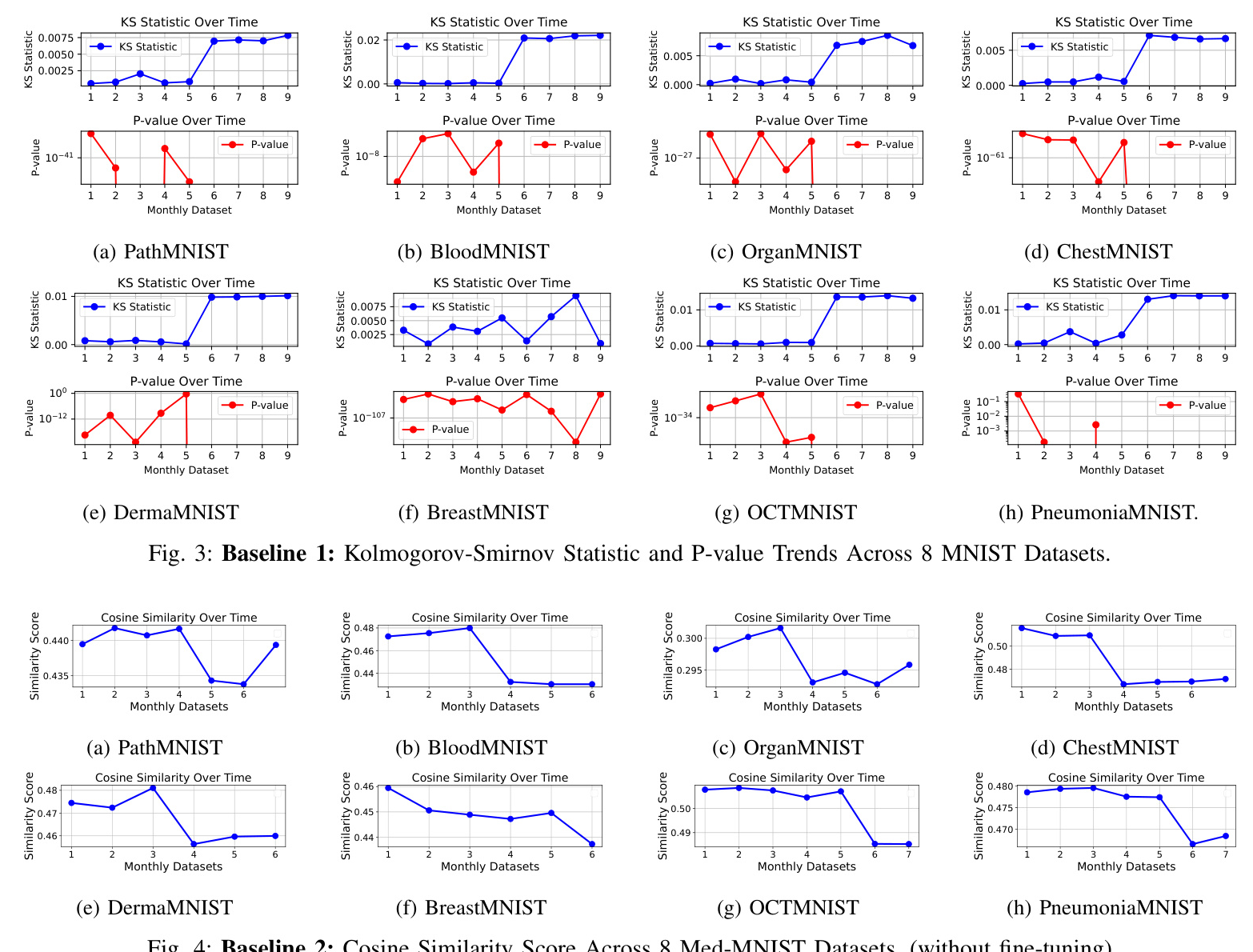
Baseline 2 (without data-sketches and fine-tuning)
In Fig. 4, it is clear that the cosine similarity scores across the 8 MedMNIST datasets remain consistently low. This suggests that the feature extraction process, without fine-tuning, performs poorly in capturing meaningful similarities between highly similar images. Despite the datasets containing images that should be visually close, the cosine similarity scores indicate otherwise.
Baseline 3 (fine-tuned without data-sketches and noises)
In Fig. 5, it is illustrated that without data-sketches, there is noticeable fluctuation in the cosine similarity scores across different datasets over time. This fluctuation indicates inconsistencies in data quality, leading to uneven similarity comparisons. The variations suggest that certain datasets have more significant differences, resulting in a drop in similarity scores.
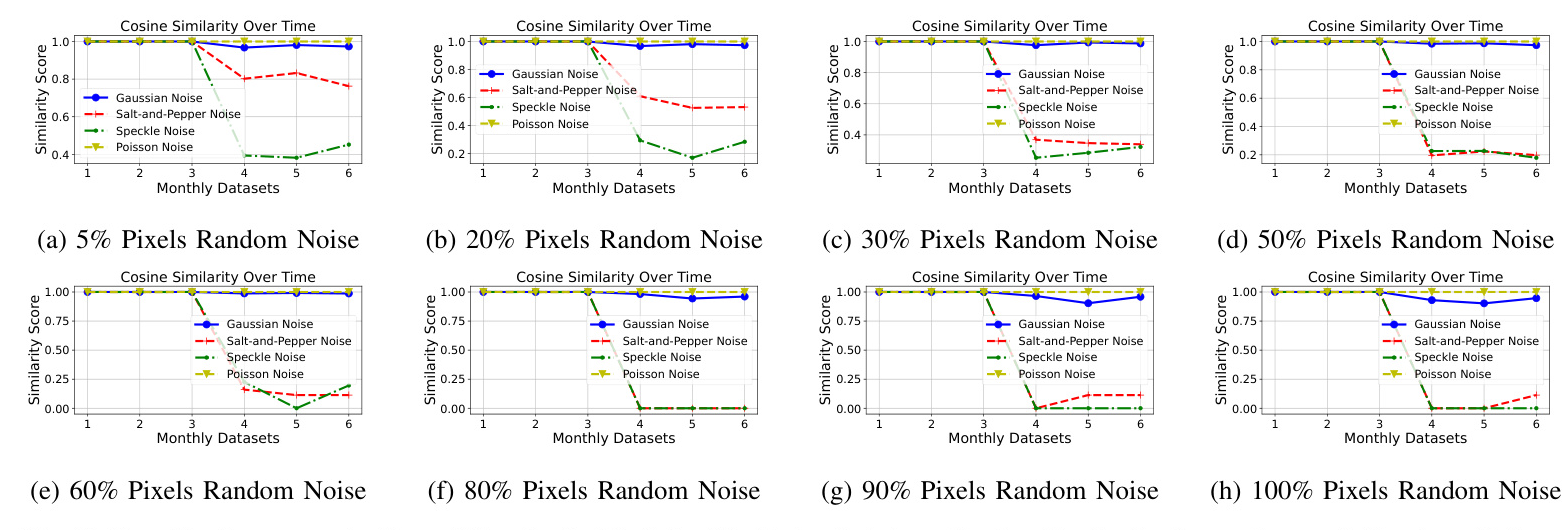
Our Performance (fine-tuned with data-sketches)
In Fig. 6, the cosine similarity scores using BreastMNIST with varying levels of noise demonstrate that the feature extraction process is of high quality. For the first three monthly datasets, the similarity scores are highly stable (100% similarity with no fluctuations) across different datasets, indicating no significant drift and consistent feature representation. This stability reflects our solution’s robustness in handling abnormal and bad quality datasets.

Sensitivity Evaluation
In the background, Section II-B, we introduced several factors that can cause dataset drift. Here, we introduce four types of noise—Gaussian Noise, Salt-and-Pepper Noise, Speckle Noise, and Poisson Noise—each corresponding to one of these drift-inducing factors. These noises are used to test the sensitivity of our method under different conditions that may simulate real-world drift scenarios.
- Gaussian Noise: Often associated with sensor performance fluctuations or low-light conditions.
- Salt-and-Pepper Noise: Linked to transmission errors or intentional tampering.
- Speckle Noise: Commonly observed in coherent imaging systems such as ultrasound, radar, or SAR.
- Poisson Noise: Related to the statistical nature of photon counting and is often linked to lighting conditions.
Fig. 7 shows the cosine similarity scores over time with just 1% random noise applied to the Med-MNIST datasets. The results highlight the sensitivity of the model, as even with this minimal noise level, the system is able to detect drift effectively.
[illustration: 7]
Related Work
Current research sought to mitigate the limitations of data drift by utilizing transfer learning, which adapts pre-trained models to new datasets with minimal training. While this method effectively reduces the data requirements, it also exposes the models to potential inaccuracies due to noise and outliers in the data. The sensitivity of deep learning models to such imperfections can significantly impair their ability to detect subtle distributional drifts, thereby undermining the reliability of diagnostic decisions.
Most of the existing research on drift detection primarily focuses on textual datasets, where distributional shifts are typically identified by monitoring changes in word embeddings or other text-based features. Drift detection in medical imaging has received relatively less attention. Unlike textual data, medical images are high-dimensional, and small, subtle changes can have significant implications. Existing techniques for drift detection in images often struggle with the complexity and high-dimensionality of image data, leading to less effective monitoring of distributional shifts. Some approaches rely on visual inspection or traditional image processing techniques, which may not be scalable or effective for real-time detection in large image datasets.
Conclusion
This paper introduces an advanced method for detecting distributional drift in CT-scan medical imaging by utilizing data-sketching techniques and fine-tuning a ViT model. The approach enhances anomaly detection accuracy and stability, offering a scalable solution for maintaining model performance in dynamic clinical settings. By leveraging data sketches and cosine similarity, the method enables efficient real-time image comparison, improving model robustness against subtle data shifts.