Authors:
Guanchu Wang、Junhao Ran、Ruixiang Tang、Chia-Yuan Chang、Chia-Yuan Chang、Yu-Neng Chuang、Zirui Liu、Vladimir Braverman、Zhandong Liu、Xia Hu
Paper:
https://arxiv.org/abs/2408.08422
Introduction
Large Language Models (LLMs) have shown remarkable capabilities in various domains, including medical research. However, their performance in diagnosing rare diseases remains uncertain. Rare diseases, despite affecting a small portion of the population, collectively impose significant public health burdens. Diagnosing these conditions is challenging due to their complex genetic origins and unpredictable clinical manifestations. This study aims to assess the diagnostic performance of LLMs in rare diseases and explore methods to enhance their effectiveness.
Preliminaries
Large Language Models for Rare Disease Diagnosis
LLMs have demonstrated competitiveness in identifying rare diseases. Previous research has shown their effectiveness in ranking causal genetic mutations and predicting patients’ phenotypes from clinical notes. However, most existing studies utilize closed-source frameworks and data, presenting challenges for reproducibility and practical deployment. This study contributes to open-source efforts by introducing the ReDis-QA dataset and the ReCOP corpus, both dedicated to rare disease question-answering.
Retrieval Augmentation Generation
Retrieval-Augmented Generation (RAG) enhances LLM performance by leveraging retrieval-based methods to supply additional knowledge for inference. A standard RAG framework comprises a corpus, retriever, and LLM. The corpus serves as a comprehensive knowledge collection, while retrievers select relevant knowledge based on the question. LLMs then generate answers using prompts based on the retrieved knowledge, providing more accurate and contextually appropriate responses. This approach is particularly powerful for tasks requiring access to extensive external knowledge, such as diagnosing rare diseases.
The Rare Disease Question Answering (ReDis-QA) Dataset
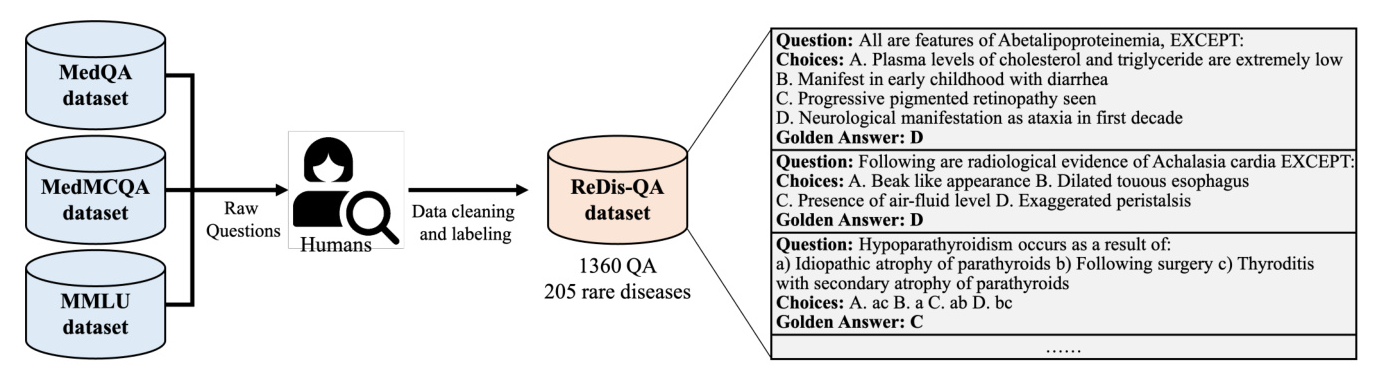
The ReDis-QA dataset is introduced to evaluate the performance of LLMs in diagnosing rare diseases. It includes 1360 high-quality question-answer pairs covering 205 rare diseases. The data collection pipeline involves data cleaning and labeling, as illustrated in Figure 1.
Data Source and Cleaning
The data sources for ReDis-QA include the MedMCQA, MedQA, and MMLU datasets. These sources provide over 200,000 raw question-answer pairs with high topical diversity. The dataset cleaning process focused on removing questions irrelevant to rare diseases, resulting in 1360 high-quality question-answer pairs relevant to rare disease diagnosis.
Data Labeling
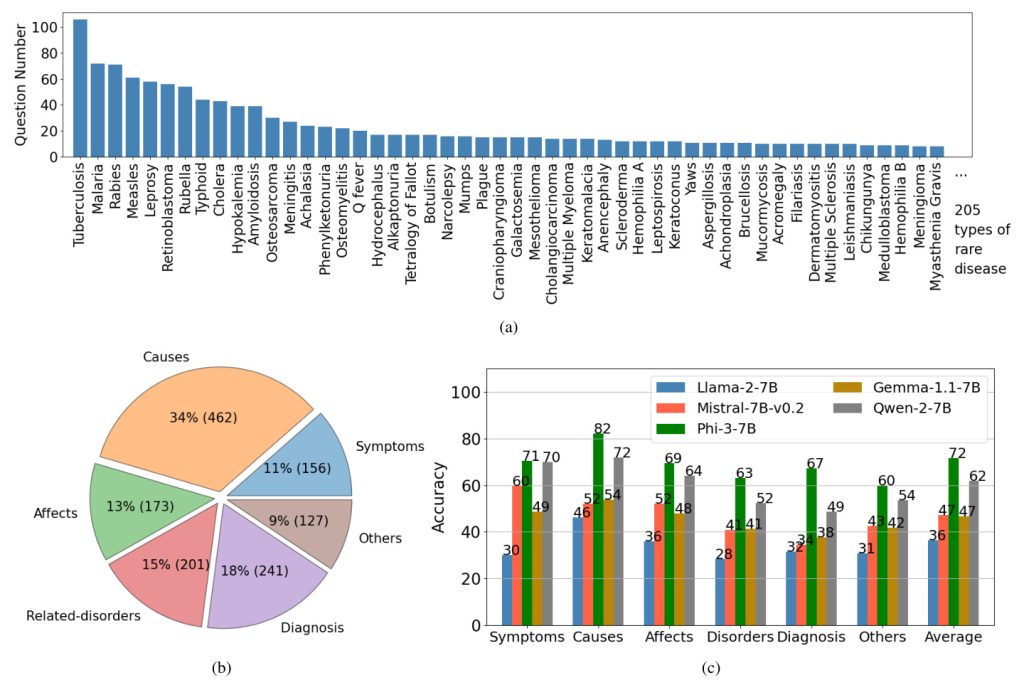
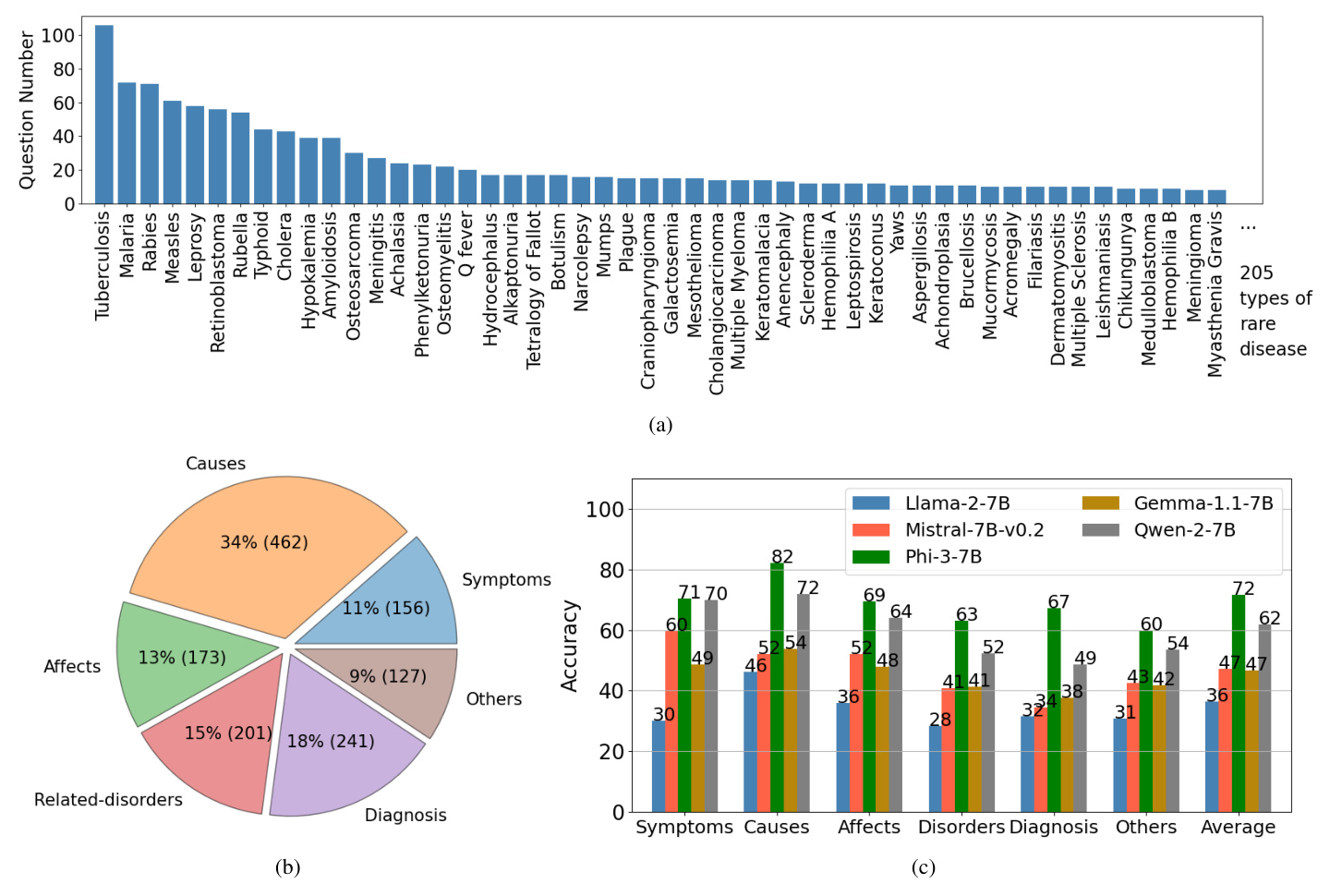
Data labeling involves annotating meta-data for each question-answer pair, including the rare disease name and property (symptoms, causes, effects, related disorders, diagnosis, or others). The statistics of the ReDis-QA dataset are shown in Figure 2. The dataset widely covers 205 types of rare diseases, with the most frequent disease featuring over 100 questions.
Benchmark of LLMs on ReDis-QA Dataset
Several open-source LLMs were benchmarked on the ReDis-QA dataset to study their capabilities in rare disease diagnosis.
Experimental Setup
The experiments were conducted using Llama-2-7B, Mistral-7B-v0.2, Phi-3-7B, Gemma-1.1-7B, and Qwen-2-7B LLMs. The evaluation metric was the accuracy on the ReDis-QA dataset.
Benchmark Results
The benchmark results, illustrated in Figure 2, reveal that diagnosing rare diseases remains a significant challenge for current open-source LLMs. Phi-3-7B showed the most competitive performance across all properties, while other models exhibited lower accuracy.
The Rare Disease Corpus
The ReCOP corpus, sourced from the National Organization for Rare Disorders (NORD) database, aims to enhance the diagnostic capabilities of LLMs for rare diseases through retrieval-augmented generation.
Data Source
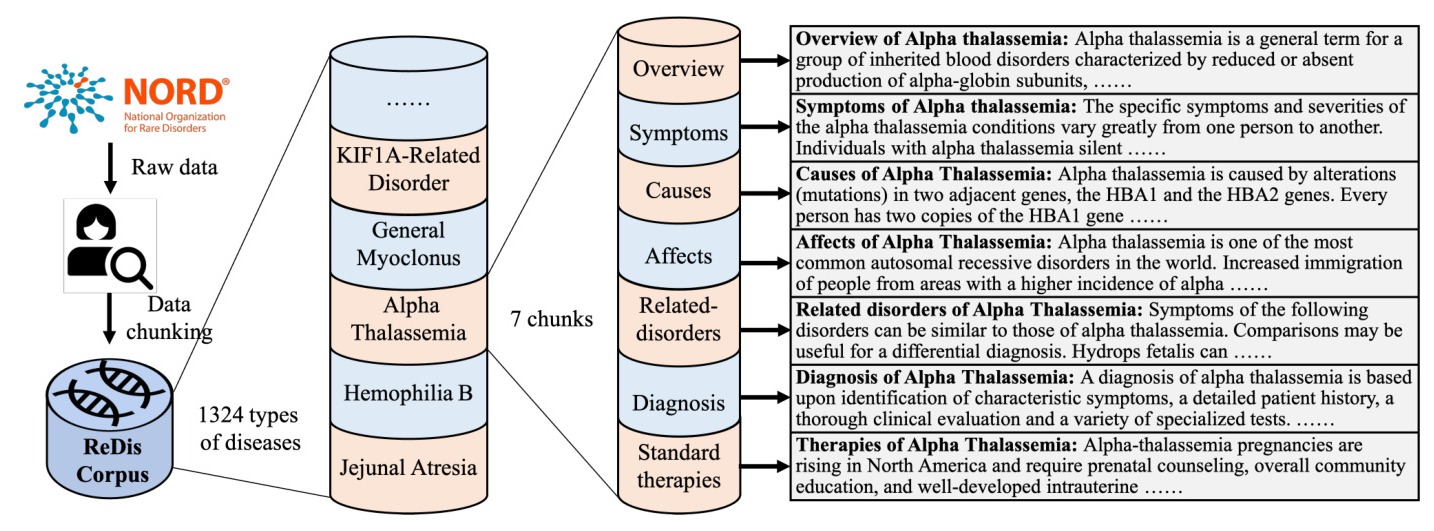
The NORD database contains professional reports on 1324 rare diseases, including comprehensive information on symptoms, causes, effects, treatments, and clinical trials. The reports are written in non-technical language, making them accessible to both non-professional individuals and LLMs.
Data Chunking
Data chunking is crucial for retrieval augmentation. Chunks are the minimal units used to match queries and provide prompts during retrieval augmentation. ReCOP divides each rare disease report into chunks: overview, symptoms, causes, effects, related disorders, diagnosis, and standard therapies. This structure ensures consistent relations between documents and queries.
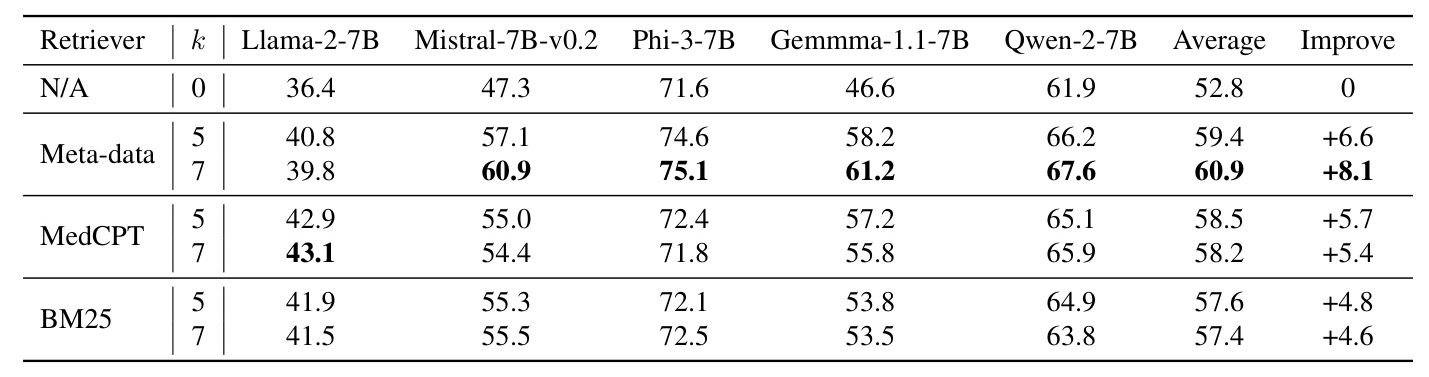
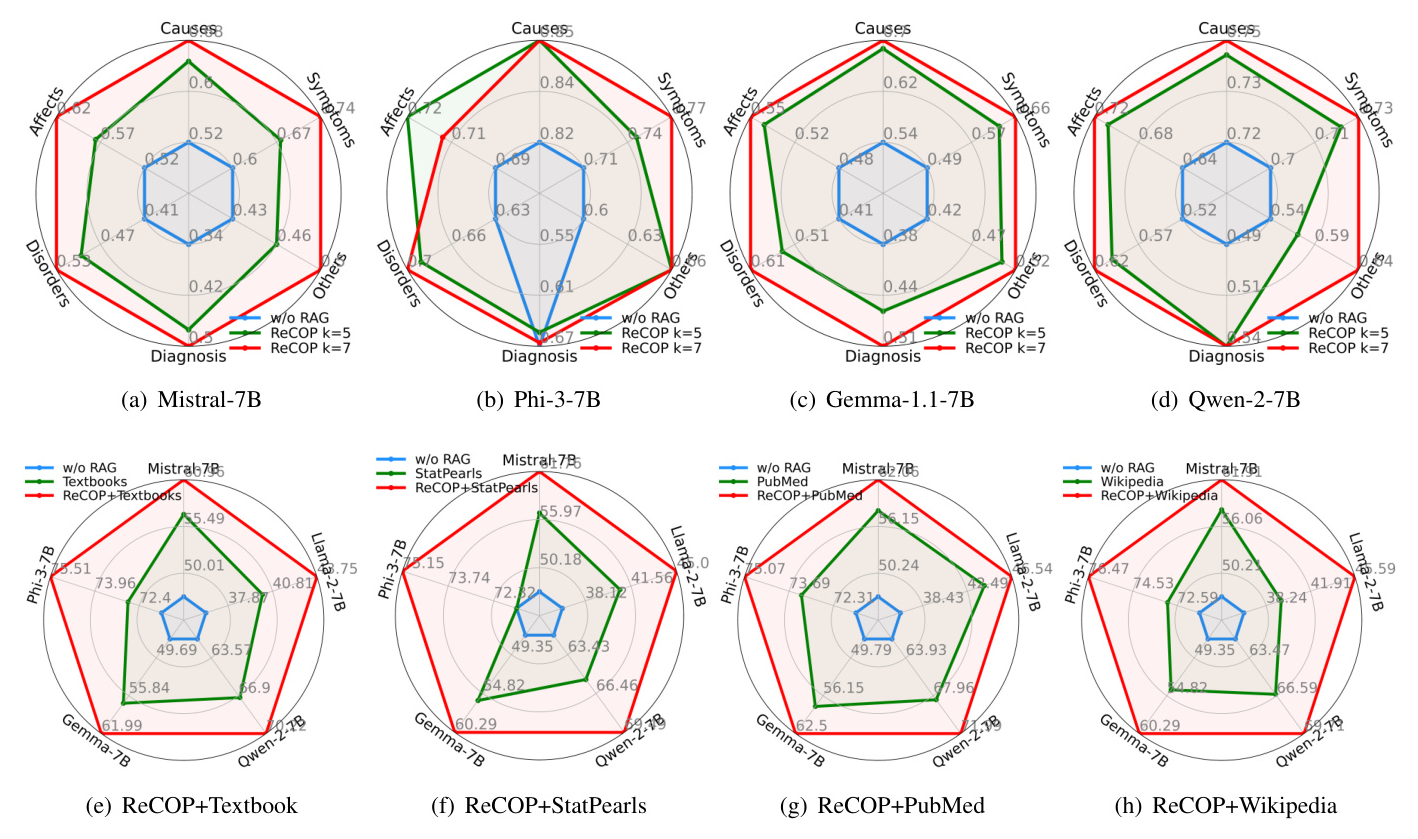
Benchmark of Retrieval Augmentation with ReCOP on ReDis-QA Dataset
Experiments demonstrate that ReCOP can significantly improve LLM performance in rare disease QA by contributing to the retrieval augmentation generation (RAG) of LLMs.
Experimental Setup
The experiments used Llama-2-7B, Mistral-7B-v0.2, Phi-3-7B, Gemma-1.1-7B, and Qwen-2-7B LLMs. Retrieval algorithms included the meta-data retriever, MedCPT (dense retriever), and BM25 (sparse retriever). Baseline corpora for comparison included PubMed, Textbook, StatPearls, and Wikipedia.
Benchmark Results
The results show that LLMs with ReCOP outperform LLMs without RAG by an average of 8%. The meta-data retriever outperformed MedCPT and BM25, indicating the usefulness of labeled meta-data and chunking in ReCOP. Combining ReCOP with other corpora significantly enriched the knowledge for LLMs, improving their diagnostic capabilities for rare diseases.
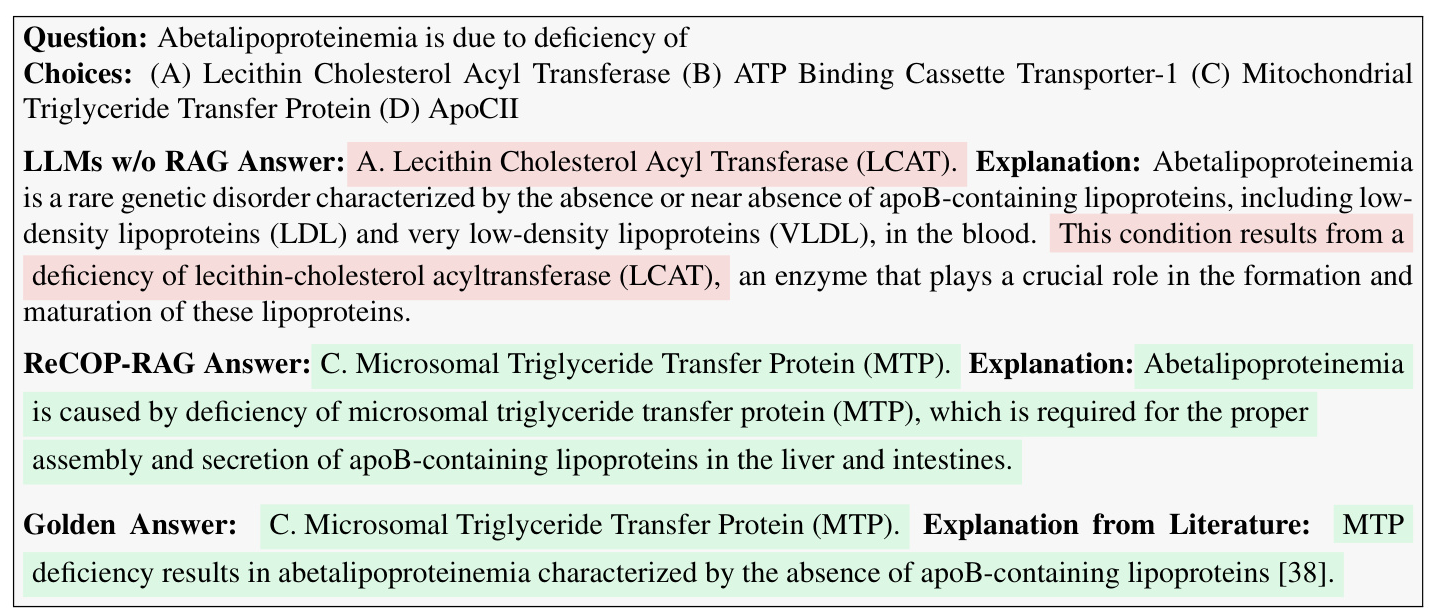
Case Studies on Natural Language Explanation
Natural language explanations are crucial for medical-related tasks. Case studies show that LLMs without RAG often follow incorrect inference processes and produce wrong answers. In contrast, ReCOP guides LLMs to the correct answer and ensures they follow the correct reasoning process, with explanations traceable to existing literature.
Conclusion
This study introduces the ReDis-QA dataset and the ReCOP corpus to benchmark and enhance the capabilities of LLMs in diagnosing rare diseases. The ReDis-QA dataset includes 1360 high-quality question-answer pairs covering 205 rare diseases, while ReCOP provides comprehensive expertise on rare diseases for retrieval-augmented generation. Experimental results demonstrate that ReCOP significantly improves the accuracy of LLMs in rare disease diagnosis and enhances their trustworthiness by providing traceable explanations.