Authors:
Anders Gjølbye、Lina Skerath、William Lehn-Schiøler、Nicolas Langer、Lars Kai Hansen
Paper:
https://arxiv.org/abs/2408.08065
Introduction
Electroencephalography (EEG) research has traditionally focused on narrowly defined tasks, but recent advancements are leveraging unlabeled data within larger models for broader applications. This shift addresses a critical challenge in EEG research: managing high noise levels in EEG data. Kostas et al. (2021) demonstrated that self-supervised learning (SSL) outperforms traditional supervised methods. However, current preprocessing methods often fail to efficiently handle the large data volumes required for SSL due to their lack of optimization and reliance on subjective manual corrections. This paper introduces SPEED, a Python-based EEG preprocessing pipeline optimized for SSL, designed to efficiently process large-scale data, stabilize self-supervised training, and enhance performance on downstream tasks.
Background
EEG data is highly variable and susceptible to noise and artifacts, necessitating robust signal cleaning methods. Historically, MATLAB has been the dominant platform for EEG research, with frameworks like EEGLAB, Brainstorm, and FieldTrip being widely used. However, these frameworks are not optimized for the massive data volumes required for SSL. Python-based tools like MNE offer comprehensive suites for EEG analysis, but they also fall short in handling large-scale data for SSL. Recent studies by Kostas et al. and Banville et al. have shown the efficacy of SSL in capturing EEG-specific properties, but they perform minimal preprocessing. This paper proposes a more comprehensive preprocessing approach to improve model performance.
Data
TUH EEG Corpus (TUEG)
The Temple University Hospital EEG Corpus is a substantial collection of clinical EEG recordings, consisting of 26,846 recordings collected from 2002 to 2017, taking up 1.6 TB of storage. This dataset is the primary data source for developing and testing the self-supervised learning pipeline.
Motor Movement/Imagery Dataset (MMIDB)
The MMIDB dataset includes 64-channel EEG recordings from 109 subjects performing motor and imagery tasks related to hand movements. This dataset is used for downstream benchmarking.
BrainCapture Bhutan Dataset v. 4.1 (BC Bhutan)
The BC Bhutan dataset includes 27-channel EEG recordings from 133 subjects across Bhutan. This dataset is used for fine-tuning models and downstream benchmarking.
BCI Challenge @ NER 2015 (BCI@NER)
The dataset from the BCI Challenge at NER 2015 includes 56-channel EEG recordings from 16 subjects participating in 5 sessions each. This dataset is used for downstream benchmarking.
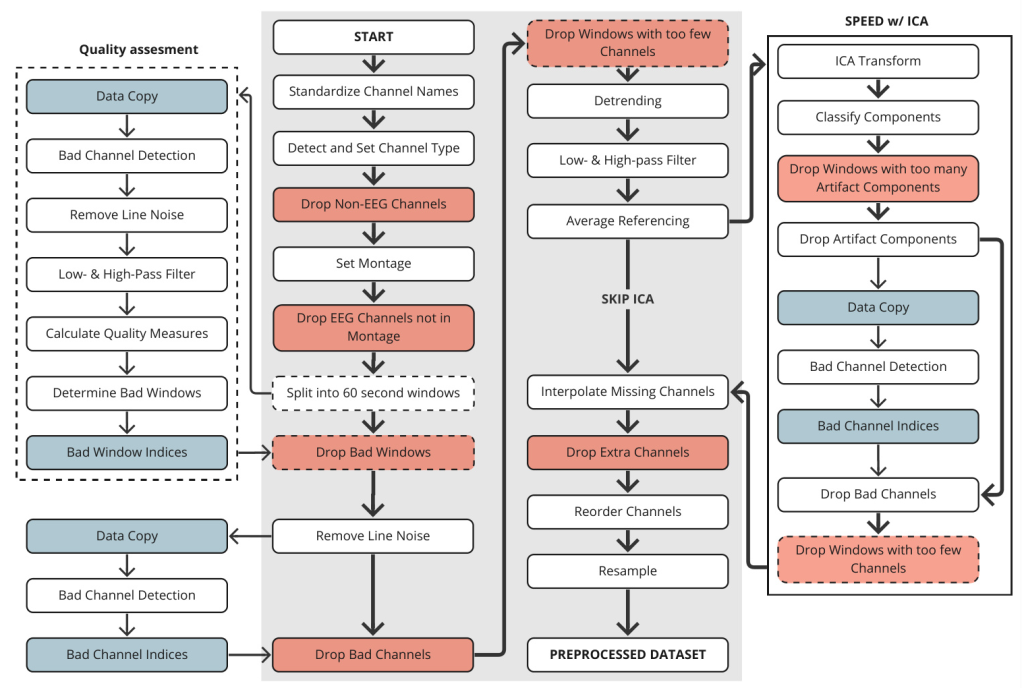
Methods
The SPEED pipeline is built upon existing preprocessing components available in various Python packages. The objective is to preprocess data into a uniform format suitable for machine learning applications, ensuring high-quality data with consistency in channels and sample counts. The general SPEED pipeline for self-supervised pretraining is described below.
Initial Setup of Preprocessing
The initial steps involve standardizing channel names, detecting channel types, dropping non-EEG channels, setting montage, and segmenting the data into 60-second windows. All subsequent procedures are carried out within these windows.
Quality Assessment
Quality assessment involves quickly determining whether a window should be kept or discarded. This step includes bad channel detection, applying a notch filter to eliminate line noise, and calculating various quality measures. Windows with poor quality are excluded based on predefined thresholds.
Iterative Zapline
The Iterative Zapline Algorithm is used to remove line noise from the data. This algorithm combines spectral and spatial filtering techniques to achieve effective noise removal while preserving the underlying signals.
Bad Channel Detection
Bad channel detection identifies channels with low signal-to-noise ratios (SNR) and removes them from the dataset. This process is essential for improving interpolation, average referencing, and independent component analysis (ICA).
Filtering & Average Referencing
Robust detrending, high-pass filtering, low-pass filtering, and average referencing are applied to the remaining high-quality channels.
Independent Component Analysis
Extended Infomax ICA transformation is performed, and components are classified using ICLabel methods. Components not categorized as brain or other with certainty above 80% are excluded. The signal is reconstructed from the remaining independent components.
Interpolation & Resample
Missing channel interpolation is performed to standardize EEG data from various sources. The channels are then reordered, and the data is resampled to 256Hz.
Downstream Data Preprocessing
Downstream data preprocessing is similar to pretraining data but with some modifications. The data is not split into 60-second windows, and quality assessment is skipped to avoid increasing the risk of Type I errors.
Experiments
Preprocessing of the TUH EEG Corpus
SPEED is designed for multiprocessing and is executed in parallel across multiple cores, completing the task in under a week. Comprehensive logs are maintained for each step, allowing detailed inspection of the data.
Pretraining
The BENDR model from Kostas et al. is pretrained using three different preprocessed versions of the TUH EEG Corpus: SPEED, SPEED w/ ICA, and Baseline. The model with the lowest validation loss is selected for subsequent probing.
Probing
Probing involves incorporating the pre-trained encoder layer from the model, adding linear layers, and locking the pre-trained encoder layer. The models are probed using the labelled downstream datasets, and a 10-fold cross-validation with subject-based splits is implemented to ensure thorough evaluation.
Results
Preprocessing
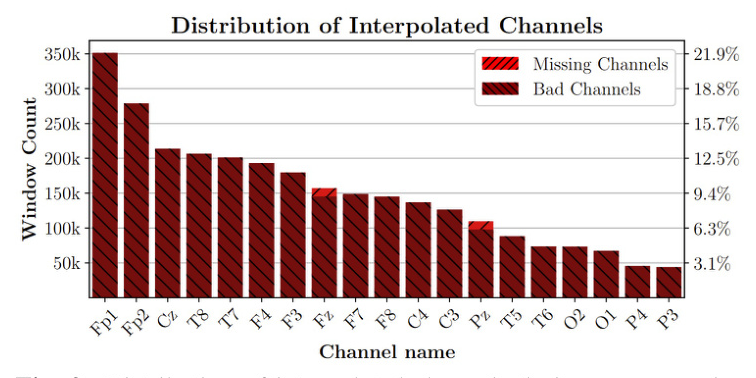
The distribution of interpolated channels during preprocessing with SPEED and SPEED w/ ICA is shown in Figure 2. Most channels are interpolated for being detected as bad, while a few instances were originally missing.
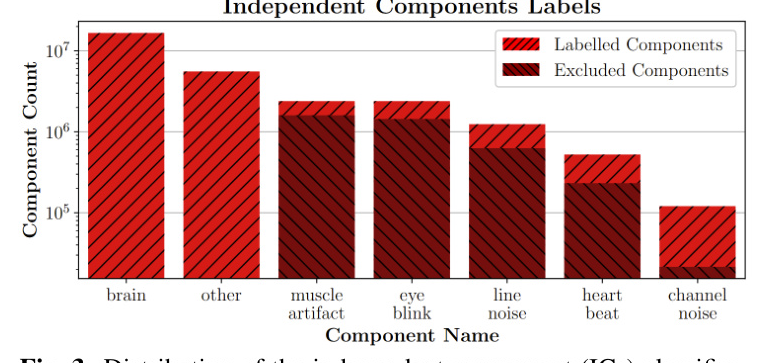
The distribution of independent component (ICs) classification by ICLabel during preprocessing with the SPEED pipeline on the TUH EEG Corpus is shown in Figure 3. Most ICs are classified as brain and other, as expected.
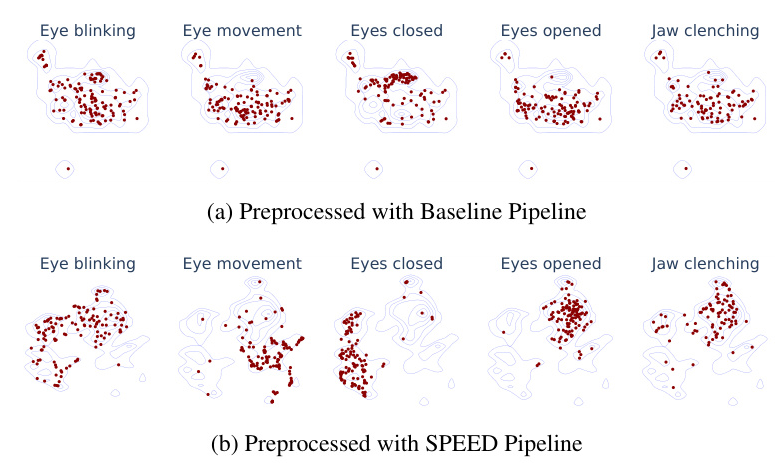
UMAP Embedding Analysis
UMAP embeddings of the Bhutan dataset show significant improvements in class separation when using the preprocessing approach. The representations based on SPEED show better alignment with the ground truth labels.
Pretraining
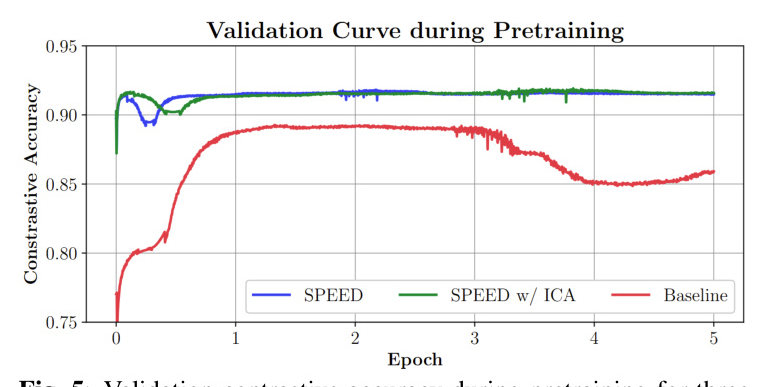
The validation contrastive accuracy curves for pretraining using three different preprocessed versions of the TUH EEG Corpus are shown in Figure 5. Models using SPEED and SPEED w/ ICA offer more stable pretraining and achieve higher contrastive accuracy compared to the Baseline model.
Probing of the Model
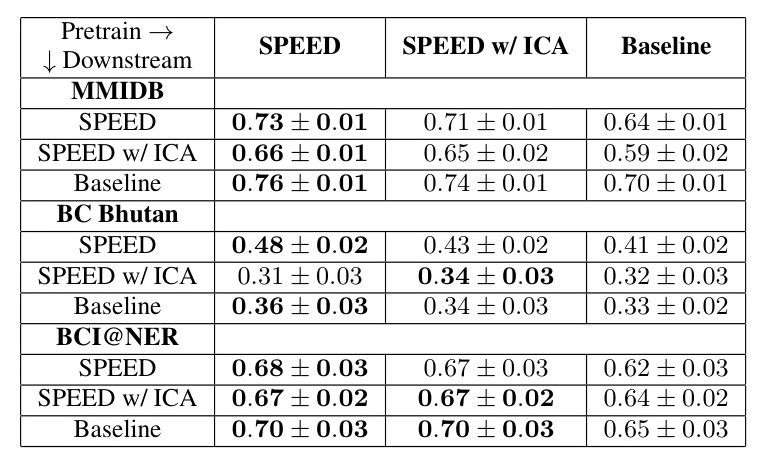
The results of probing the model are presented in Table 1. Both SPEED and SPEED w/ ICA during pretraining enhance the model’s downstream accuracy compared to the baseline. However, SPEED w/ ICA does not outperform SPEED.
Discussion
The SPEED pipeline addresses a major challenge of large-scale EEG data preprocessing. However, the inclusion of ICA with ICLabel does not significantly improve pretraining performance and may slightly reduce downstream accuracy. For downstream datasets, more careful preprocessing is often required. The potential bias introduced by fine-tuning the model using the same datasets designated for validation during pretraining is not a major concern due to the use of 60-second windows and all available data.
Conclusion
The SPEED pipeline improves stability, convergence, and contrastive accuracy during pretraining and produces a more suitable latent space for downstream classification tasks. The probing results show significant improvement for several downstream classification tasks when pretraining using the SPEED pipeline compared to a simple baseline preprocessing pipeline. The tools for reproducible preprocessing of the TUH EEG Corpus are ready for future development of pretrained EEG foundation models. The results provide evidence that physically motivated preprocessing is useful for self-supervised learning of EEG representations.