Authors:
Xiaomin Wu、Rui Xu、Pengchen Wei、Wenkang Qin、Peixiang Huang、Ziheng Li、Lin Luo
Paper:
https://arxiv.org/abs/2408.07037
Introduction
Pathological examination is the gold standard for diagnosing tumors and cancers. The meticulous analysis of tissue samples by experienced pathologists provides critical insights into appropriate therapeutic strategies. However, the scarcity of senior pathologists and the cumbersome processes of accessing and consulting pathological knowledge exacerbate the complexity and workload inherent in the diagnostic process.
The advent of digital tools like Whole Slide Imaging (WSI) has facilitated pathological diagnosis, making data storage and transfer easier. Concurrently, large language models (LLMs) are increasingly utilized for their capabilities in machine learning and deep learning algorithms, particularly in analyzing pathological images. Despite these advancements, the efficacy of these models is often limited by the availability of large annotated datasets and their specialized application domains.
To bridge this gap, we propose a comprehensive, user-friendly multimodal model that integrates essential capabilities for pathologists and medical students. This model encompasses image analysis, pathology-related image queries, retrieval of pathological knowledge, and the generation of complete diagnostic reports.
Methods
PathEnhanceDS Compiling
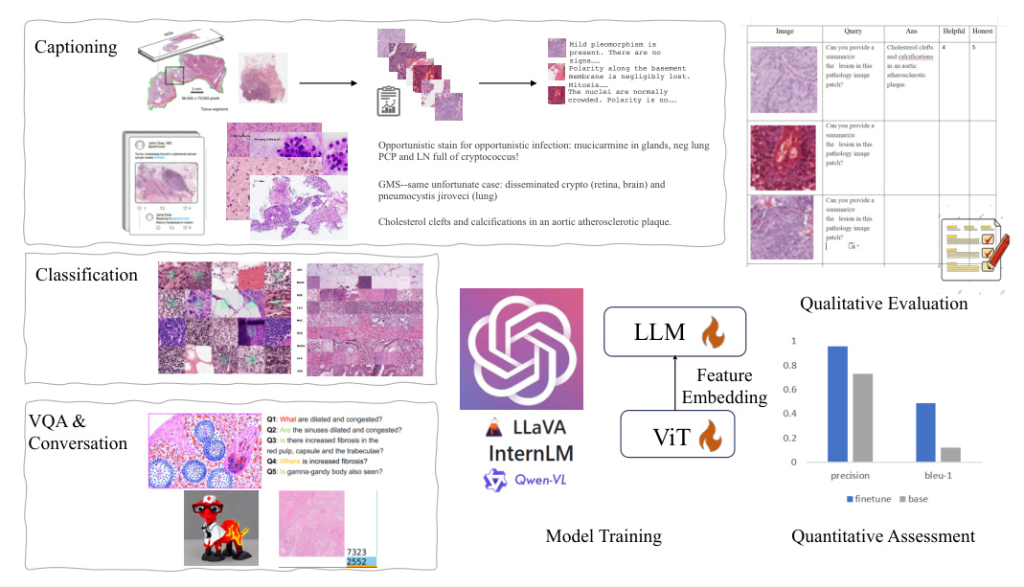
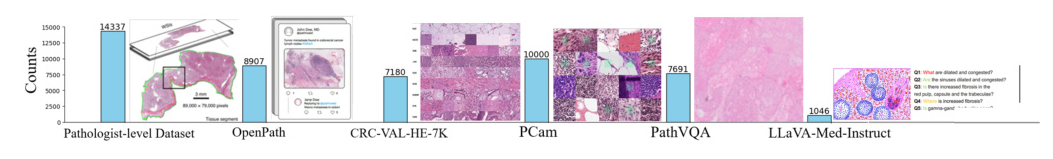
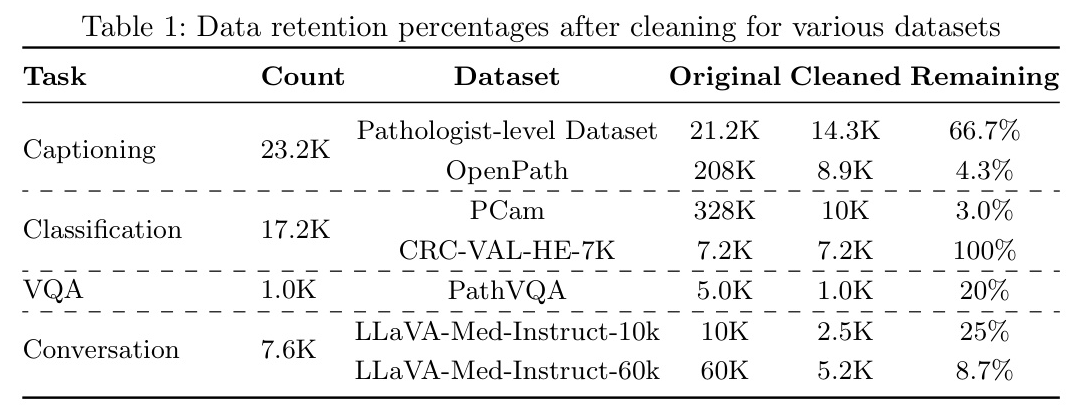
To ensure the comprehensiveness and diversity of our dataset, we meticulously curated data from various sources, resulting in the creation of PathEnhanceDS. This dataset includes approximately 45,000 instances covering tasks such as pathological image grading, classification, caption generation, image descriptions, image-based question-answering, and dialogue interactions.
We selected datasets based on their relevance to diagnostic pathology, quality of annotations, and compatibility across different tasks. Key datasets include:
- Pathologist-level Dataset: Annotated by medical professionals, providing patch-level descriptive information.
- OpenPath: Derived from Twitter, offering unconventional perspectives on pathology data.
- PCam: Recognized for its binary classification challenges within the pathology domain.
- CRC-VAL-HE-7K: Focused on multi-classification of tissue organs.
- PathVQA: Comprehensive compilation of pathology images paired with question-answer pairs.
- LLaVA-Med-Instruct: Characterized by conversation-based queries that replicate diagnostic thought processes.
The arrangement of data forms a single-round interaction in the format:
– User: <img>Pv</img> Pq<STOP>
– Assistant: Pd<STOP>
This diverse dataset construction was instrumental in creating an effective and wide-reaching study.
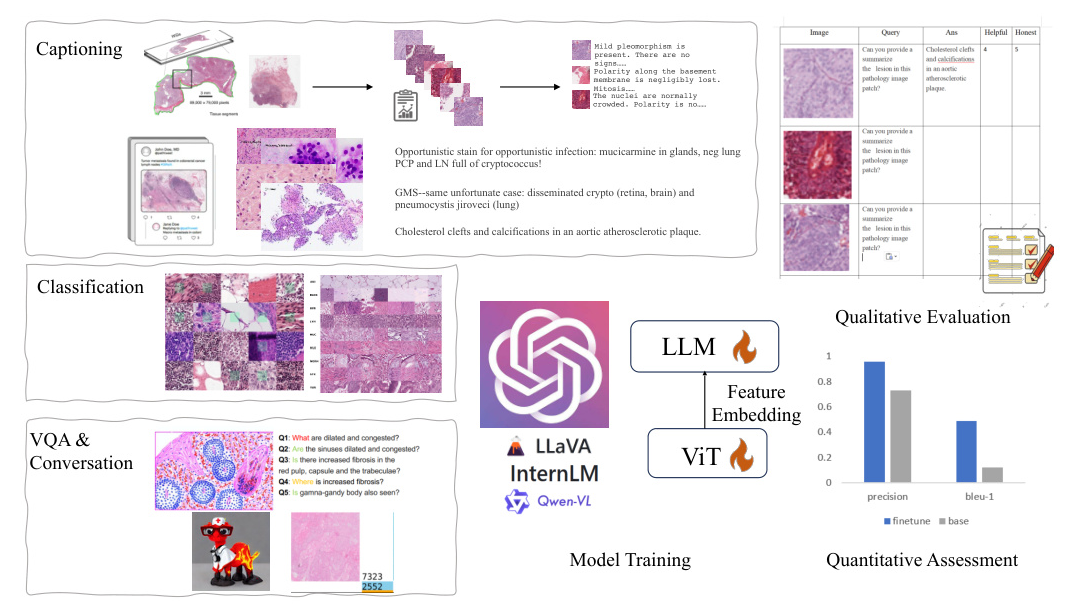
PathoSync Tuning
We experimented with three representative multimodal large language models: LLaVA, Qwen-VL, and InternLM. These models, recognized for their performance in natural visual language domains, were fine-tuned using our PathEnhanceDS dataset.
- LLaVA: Based on the LLaMA language model, employing a vision encoder to convert images into patch features.
- Qwen-VL-7B: Integrates the ViT-bigG project as its vision encoder, facilitating close interaction between textual and visual information.
- InternLM: A groundbreaking vision-language large model excelling in free-form text-image composition and comprehension.
To bridge the gap between general-purpose models and the medical histopathology domain, we fully enabled the fine-tuning of the Vision Transformer (ViT) component during training. We experimented with both full-parameter tuning and Low-Rank Adaptation (LoRA) training methods.
The training process involved setting the sequence length to 2048, encoding text using a tokenizer, and extracting image paths accordingly. The learning rate was set to 1e-5, batch size to 2, and the warm-up ratio to 0.05. The model was trained on 45,000+ data points for 3 epochs, with checkpoints saved for subsequent evaluation.
Experiments
Evaluation Metrics
To verify the effectiveness of fine-tuning, we focused on core tasks such as classification of pathological images, caption generation, and visual reasoning based on pathological imagery. Evaluation metrics included precision, recall, and F1 score for classification tasks, and BLEU and ROUGE scores for open-ended generative tasks.
Evaluation Results
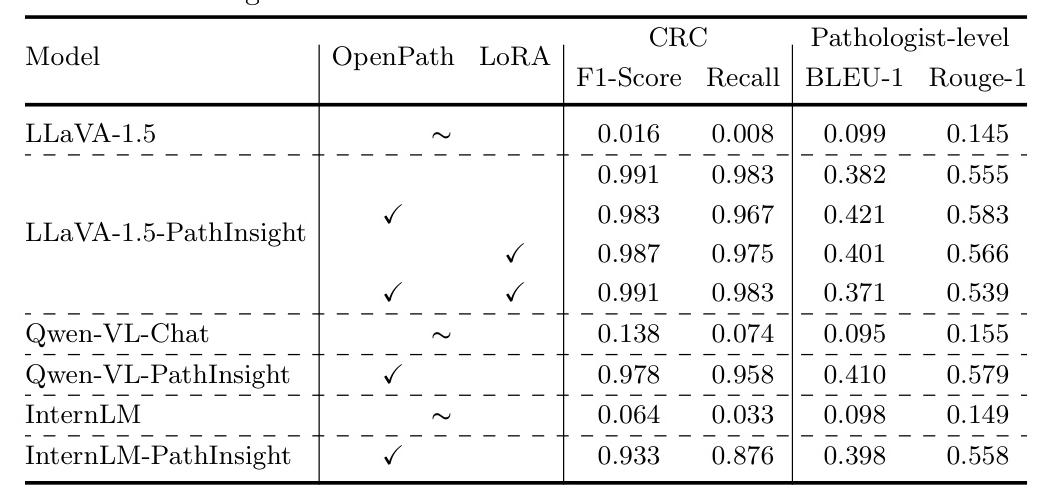
The results indicated significant improvements in model performance after fine-tuning. For CRC classification tasks, the model’s performance was highly consistent with the ground truth. The generated captions were structurally accurate and generally conformed to fundamental medical pathology knowledge, though some discrepancies in detail accuracy remained.
Quantitative results showed improvements in precision, recall, and F1 scores for classification tasks. For example, the F1-score in CRC classification tasks for the Qwen-VL model increased from 0.138 to about 0.978. The LLaVA model’s recall improved from 0.008 to 0.967.
In the captioning task, the Qwen-VL model’s BLEU-1 score improved from 0.09 to 0.41, and the ROUGE-1 score improved from 0.15 to 0.58. The LLaVA-1.5 model’s BLEU-1 score improved from 0.099 to 0.421 (full parameters fine-tuning) and 0.371 (LoRA parameters fine-tuning), with corresponding improvements in ROUGE-1 scores.
Conclusion
The integration of multimodal large models has significantly improved the handling of diverse tasks in a unified framework, enhancing both image and text analysis. In computational pathology, these models enable clinicians and medical students to access and document diagnostic information more efficiently.
To address the challenges of high data acquisition costs and the scarcity of quality datasets, we developed PathEnhanceDS, a comprehensive dataset with over 45,000 cases. By fine-tuning models like Qwen-VL, InternLM, and LLaVA, our evaluations show significant performance improvements in pathology, adhering to clinical medicine standards for open-ended questions. This highlights the effectiveness of our fine-tuning approach and the adaptability of our carefully curated dataset.